PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca2+ homeostasis
- PMID: 30892128
- PMCID: PMC6844518
- DOI: 10.1080/15548627.2019.1596489
PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca2+ homeostasis
Abstract
PSEN2 (presenilin 2) is one of the 3 proteins that, when mutated, causes early onset familial Alzheimer disease (FAD) cases. In addition to its well-known role within the γ-secretase complex (the enzyme ultimately responsible for Aβ peptides formation), PSEN2 is endowed with some γ-secretase-independent functions in distinct cell signaling pathways, such as the modulation of intracellular Ca2+ homeostasis. Here, by using different FAD-PSEN2 cell models, we demonstrate that mutated PSEN2 impairs autophagy by causing a block in the degradative flux at the level of the autophagosome-lysosome fusion step. The defect does not depend on an altered lysosomal functionality but rather on a decreased recruitment of the small GTPase RAB7 to autophagosomes, a key event for normal autophagy progression. Importantly, FAD-PSEN2 action on autophagy is unrelated to its γ-secretase activity but depends on its previously reported ability to partially deplete ER Ca2+ content, thus reducing cytosolic Ca2+ response upon IP3-linked cell stimulations. Our data sustain the pivotal role for Ca2+ signaling in autophagy and reveal a novel mechanism by which FAD-linked presenilins alter the degradative process, reinforcing the view of a causative role for a dysfunctional quality control pathway in AD neurodegeneration.Abbreviations: Aβ: amyloid β; AD: Alzheimer disease; ACTB: actin beta; AMPK: AMP-activated protein kinase; APP: amyloid-beta precursor protein; BafA: bafilomycin A1; BAPTA-AM: 1,2-bis(o-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid acetoxymethyl ester; CFP: cyan fluorescent protein; EGTA-AM: ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid acetoxymethyl ester; ER: endoplasmic reticulum; EGFP-HDQ74: enhanced GFP-huntingtin exon 1 containing 74 polyglutamine repeats; FAD: familial Alzheimer disease; FCS: fetal calf serum; FRET: fluorescence/Förster resonance energy transfer; GFP: green fluorescent protein; IP3: inositol trisphosphate; KD: knockdown; LAMP1: lysosomal associated membrane protein 1; MAP1LC3-II/LC3-II: lipidated microtubule-associated protein 1 light chain 3; MCU: mitochondrial calcium uniporter; MICU1: mitochondrial calcium uptake 1; MEFs: mouse embryonic fibroblasts; MFN2: mitofusin 2; MTOR: mechanistic target of rapamycin kinase; MTORC1: MTOR complex 1; SQSTM1/p62: sequestosome 1; PSEN1: presenilin 1; PSEN2: presenilin 2; RAB7: RAB7A: member RAS oncogene family; RFP: red fluorescent protein; ATP2A/SERCA: ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting; siRNA: small interference RNA; V-ATPase: vacuolar-type H+-ATPase; WT: wild type.
Keywords: ATP2A/SERCA; Alzheimer disease; ER-mitochondria tethering; RAB7; autophagosome-lysosome fusion; calcium; presenilin.
Figures
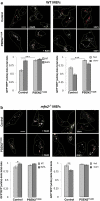
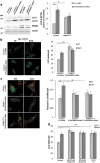
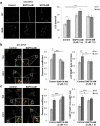
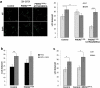
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
