Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy
- PMID: 30892132
- PMCID: PMC6601540
- DOI: 10.1080/19420862.2019.1596513
Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy
Abstract
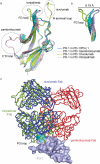
Monoclonal antibody (mAb)-based blockade of programmed cell death 1 (PD-1) or its ligand to enable antitumor T-cell immunity has been successful in treating multiple tumors. However, the structural basis of the binding mechanisms of the mAbs and PD-1 and the effects of glycosylation of PD-1 on mAb interaction are not well understood. Here, we report the complex structure of PD-1 with toripalimab, a mAb that is approved by China National Medical Products Administration as a second-line treatment for melanoma and is under multiple Phase 1-Phase 3 clinical trials in both China and the US. Our analysis reveals that toripalimab mainly binds to the FG loop of PD-1 with an unconventionally long complementarity-determining region 3 loop of the heavy chain, which is distinct from the known binding epitopes of anti-PD-1 mAbs with structural evidences. The glycan modifications of PD-1 could be observed in three potential N-linked glycosylation sites, while no substantial influences were detected to the binding of toripalimab. These findings benefit our understanding of the binding mechanisms of toripalimab to PD-1 and shed light for future development of biologics targeting PD-1. Atomic coordinates have been deposited in the Protein Data Bank under accession code 6JBT.
Keywords: PD-1; Toripalimab; complex structure; glycosylation.
Figures


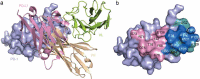

References
-
- Tan S, Gao GF.. New hope for cancer treatment: cancer immunotherapy. Chinese Sci Bull. 2015;60:3155–57. In Chinese.
-
- Nishimura H, Nose M, Hiai H, Minato N and Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
