Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing
- PMID: 30893341
- PMCID: PMC6443194
- DOI: 10.1371/journal.pgen.1007810
Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing
Abstract
Spermatogenesis is the process by which male gametes are formed from a self-renewing population of spermatogonial stem cells (SSCs) residing in the testis. SSCs represent less than 1% of the total testicular cell population in adults, but must achieve a stable balance between self-renewal and differentiation. Once differentiation has occurred, the newly formed and highly proliferative spermatogonia must then enter the meiotic program in which DNA content is doubled, then halved twice to create haploid gametes. While much is known about the critical cellular processes that take place during the specialized cell division that is meiosis, much less is known about how the spermatocytes in the "first-wave" in juveniles compare to those that contribute to long-term, "steady-state" spermatogenesis in adults. Given the strictly-defined developmental process of spermatogenesis, this study explored the transcriptional profiles of developmental cell stages during testis maturation. Using a combination of comprehensive germ cell sampling with high-resolution, single-cell-mRNA-sequencing, we have generated a reference dataset of germ cell gene expression. We show that discrete developmental stages of spermatogenesis possess significant differences in the transcriptional profiles from neonates compared to juveniles and adults. Importantly, these gene expression dynamics are also reflected at the protein level in their respective cell types. We also show differential utilization of many biological pathways with age in both spermatogonia and spermatocytes, demonstrating significantly different underlying gene regulatory programs in these cell types over the course of testis development and spermatogenic waves. This dataset represents the first unbiased sampling of spermatogonia and spermatocytes during testis maturation, at high-resolution, single-cell depth. Not only does this analysis reveal previously unknown transcriptional dynamics of a highly transitional cell population, it has also begun to reveal critical differences in biological pathway utilization in developing spermatogonia and spermatocytes, including response to DNA damage and double-strand breaks.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









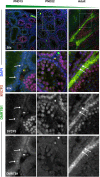
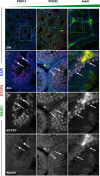


References
-
- Lawson KA, Hage WJ. Germline development. Ciba Foundation Symposium 182. Wiley; 1994.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

