Ebola Virus Entry: From Molecular Characterization to Drug Discovery
- PMID: 30893774
- PMCID: PMC6466262
- DOI: 10.3390/v11030274
Ebola Virus Entry: From Molecular Characterization to Drug Discovery
Abstract
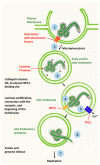
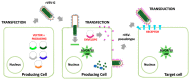
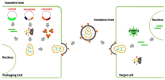
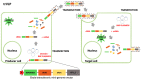
Ebola Virus Disease (EVD) is one of the most lethal transmissible infections, characterized by a high fatality rate, and caused by a member of the Filoviridae family. The recent large outbreak of EVD in Western Africa (2013⁻2016) highlighted the worldwide threat represented by the disease and its impact on global public health and the economy. The development of highly needed anti-Ebola virus antivirals has been so far hampered by the shortage of tools to study their life cycle in vitro, allowing to screen for potential active compounds outside a biosafety level-4 (BSL-4) containment. Importantly, the development of surrogate models to study Ebola virus entry in a BSL-2 setting, such as viral pseudotypes and Ebola virus-like particles, tremendously boosted both our knowledge of the viral life cycle and the identification of promising antiviral compounds interfering with viral entry. In this context, the combination of such surrogate systems with large-scale small molecule compounds and haploid genetic screenings, as well as rational drug design and drug repurposing approaches will prove priceless in our quest for the development of a treatment for EVD.
Keywords: Ebola virus; Filoviridae; VSV; antivirals; pseudovirus; retroviral vectors; small molecules; viral entry; virus-like particles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Goldstein T., Anthony S.J., Gbakima A., Bird B.H., Bangura J., Tremeau-Bravard A., Belaganahalli M.N., Wells H.L., Dhanota J.K., Liang E., et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 2018;3:1084–1089. doi: 10.1038/s41564-018-0227-2. - DOI - PMC - PubMed
-
- Subissi L., Keita M., Mesfin S., Rezza G., Diallo B., Van Gucht S., Musa E.O., Yoti Z., Keita S., Djingarey M.H., et al. Ebola Virus Transmission Caused by Persistently Infected Survivors of the 2014–2016 Outbreak in West Africa. J. Infect. Dis. 2018;218:S287–S291. doi: 10.1093/infdis/jiy280. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

