Association of Human FOS Promoter Variants with the Occurrence of Knee-Osteoarthritis in a Case Control Association Study
- PMID: 30893847
- PMCID: PMC6471183
- DOI: 10.3390/ijms20061382
Association of Human FOS Promoter Variants with the Occurrence of Knee-Osteoarthritis in a Case Control Association Study
Abstract
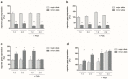
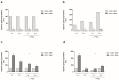
Our aim was to analyse (i) the presence of single nucleotide polymorphisms (SNPs) in the JUN and FOS core promoters in patients with rheumatoid arthritis (RA), knee-osteoarthritis (OA), and normal controls (NC); (ii) their functional influence on JUN/FOS transcription levels; and (iii) their associations with the occurrence of RA or knee-OA. JUN and FOS promoter SNPs were identified in an initial screening population using the Non-Isotopic RNase Cleavage Assay (NIRCA); their functional influence was analysed using reporter gene assays. Genotyping was done in RA (n = 298), knee-OA (n = 277), and NC (n = 484) samples. For replication, significant associations were validated in a Finnish cohort (OA: n = 72, NC: n = 548). Initially, two SNPs were detected in the JUN promoter and two additional SNPs in the FOS promoter in perfect linkage disequilibrium (LD). JUN promoter SNP rs4647009 caused significant downregulation of reporter gene expression, whereas reporter gene expression was significantly upregulated in the presence of the FOS promoter SNPs. The homozygous genotype of FOS promoter SNPs showed an association with the susceptibility for knee-OA (odds ratio (OR) 2.12, 95% confidence interval (CI) 1.2⁻3.7, p = 0.0086). This association was successfully replicated in the Finnish Health 2000 study cohort (allelic OR 1.72, 95% CI 1.2⁻2.5, p = 0.006). FOS Promoter variants may represent relevant susceptibility markers for knee-OA.
Keywords: AP-1; FOS; JUN; knee osteoarthritis; promoter.
Conflict of interest statement
The authors declare no conflicts of interests.
Figures

Similar articles
-
FOXP3 rs3761548 polymorphism is associated with knee osteoarthritis in a Turkish population.Int J Rheum Dis. 2018 Oct;21(10):1779-1786. doi: 10.1111/1756-185X.13337. Epub 2018 Aug 30. Int J Rheum Dis. 2018. PMID: 30168273
-
The single-nucleotide polymorphism (SNP) of tumor necrosis factor α -308G/A gene is associated with early-onset primary knee osteoarthritis in an Egyptian female population.Clin Rheumatol. 2017 Nov;36(11):2525-2530. doi: 10.1007/s10067-017-3727-1. Epub 2017 Jul 10. Clin Rheumatol. 2017. PMID: 28695434
-
Association of OPG gene polymorphisms with the risk of knee osteoarthritis among Chinese people.Mol Genet Genomic Med. 2019 Jun;7(6):e662. doi: 10.1002/mgg3.662. Epub 2019 Apr 11. Mol Genet Genomic Med. 2019. PMID: 30972968 Free PMC article.
-
A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis.Osteoarthritis Cartilage. 2010 May;18(5):699-704. doi: 10.1016/j.joca.2009.12.012. Epub 2010 Feb 6. Osteoarthritis Cartilage. 2010. PMID: 20175976
-
Association of Matrix Metalloproteinase (MMP) Gene Polymorphisms With Knee Osteoarthritis: A Review of the Literature.Cureus. 2021 Oct 8;13(10):e18607. doi: 10.7759/cureus.18607. eCollection 2021 Oct. Cureus. 2021. PMID: 34765365 Free PMC article. Review.
Cited by
-
Tranexamic Acid Is Beneficial to Patients Undergoing Open-Wedge High Tibial Osteotomy.Biomed Res Int. 2020 Nov 4;2020:2514207. doi: 10.1155/2020/2514207. eCollection 2020. Biomed Res Int. 2020. PMID: 33204689 Free PMC article.
-
Metabolic rewiring controlled by c-Fos governs cartilage integrity in osteoarthritis.Ann Rheum Dis. 2023 Sep;82(9):1227-1239. doi: 10.1136/ard-2023-224002. Epub 2023 Jun 21. Ann Rheum Dis. 2023. PMID: 37344157 Free PMC article.
-
Gene-Based Testing of Interactions Using XGBoost in Genome-Wide Association Studies.Front Cell Dev Biol. 2021 Dec 16;9:801113. doi: 10.3389/fcell.2021.801113. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34977040 Free PMC article.
-
Identification and validation of hub genes and potential drugs involved in osteoarthritis through bioinformatics analysis.Front Genet. 2023 Feb 9;14:1117713. doi: 10.3389/fgene.2023.1117713. eCollection 2023. Front Genet. 2023. PMID: 36845391 Free PMC article.
-
Systematic Postoperative Assessment of a Minimally-Invasive Sheep Model for the Treatment of Osteochondral Defects.Life (Basel). 2020 Dec 7;10(12):332. doi: 10.3390/life10120332. Life (Basel). 2020. PMID: 33297497 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

