Characterization of the Filovirus-Resistant Cell Line SH-SY5Y Reveals Redundant Role of Cell Surface Entry Factors
- PMID: 30893855
- PMCID: PMC6466046
- DOI: 10.3390/v11030275
Characterization of the Filovirus-Resistant Cell Line SH-SY5Y Reveals Redundant Role of Cell Surface Entry Factors
Abstract
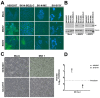
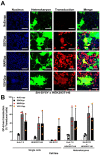
Filoviruses infect a wide range of cell types with the exception of lymphocytes. The intracellular proteins cathepsin B and L, two-pore channel 1 and 2, and bona fide receptor Niemann⁻Pick Disease C1 (NPC1) are essential for the endosomal phase of cell entry. However, earlier steps of filoviral infection remain poorly characterized. Numerous plasma membrane proteins have been implicated in attachment but it is still unclear which ones are sufficient for productive entry. To define a minimal set of host factors required for filoviral glycoprotein-driven cell entry, we screened twelve cell lines and identified the nonlymphocytic cell line SH-SY5Y to be specifically resistant to filovirus infection. Heterokaryons of SH-SY5Y cells fused to susceptible cells were susceptible to filoviruses, indicating that SH-SY5Y cells do not express a restriction factor but lack an enabling factor critical for filovirus entry. However, all tested cell lines expressed functional intracellular factors. Global gene expression profiling of known cell surface entry factors and protein expression levels of analyzed attachment factors did not reveal any correlation between susceptibility and expression of a specific host factor. Using binding assays with recombinant filovirus glycoprotein, we identified cell attachment as the step impaired in filovirus entry in SH-SY5Y cells. Individual overexpression of attachment factors T-cell immunoglobulin and mucin domain 1 (TIM-1), Axl, Mer, or dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) rendered SH-SY5Y cells susceptible to filovirus glycoprotein-driven transduction. Our study reveals that a lack of attachment factors limits filovirus entry and provides direct experimental support for a model of filoviral cell attachment where host factor usage at the cell surface is highly promiscuous.
Keywords: Filovirus cell entry; SH-SY5Y cell line; attachment factors redundancy; host–pathogen interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T., et al. Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. - DOI - PMC - PubMed
-
- Siegert R., Shu H.L., Slenczka H.L., Peters D., Muller G. The aetiology of an unknown human infection transmitted by monkeys (preliminary communication) Ger. Med. Mon. 1968;13:1–2. - PubMed
-
- CDC Outbreaks Chronology: Ebola Virus Disease|Ebola Hemorrhagic Fever|CDC. [(accessed on 13 August 2018)];2018 Available online: https://www.cdc.gov/vhf/ebola/history/chronology.html.Cdc-Ebola.
-
- CDC Outbreaks Chronology: Marburg Hemorrhagic Fever. [(accessed on 13 August 2018)];2018 Available online: https://www.cdc.gov/vhf/marburg/outbreaks/chronology.html.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

