A Temporal Order in 5'- and 3'- Processing of Eukaryotic tRNAHis
- PMID: 30893886
- PMCID: PMC6470698
- DOI: 10.3390/ijms20061384
A Temporal Order in 5'- and 3'- Processing of Eukaryotic tRNAHis
Abstract
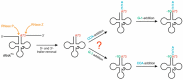
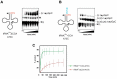
For flawless translation of mRNA sequence into protein, tRNAs must undergo a series of essential maturation steps to be properly recognized and aminoacylated by aminoacyl-tRNA synthetase, and subsequently utilized by the ribosome. While all tRNAs carry a 3'-terminal CCA sequence that includes the site of aminoacylation, the additional 5'-G-1 position is a unique feature of most histidine tRNA species, serving as an identity element for the corresponding synthetase. In eukaryotes including yeast, both 3'-CCA and 5'-G-1 are added post-transcriptionally by tRNA nucleotidyltransferase and tRNAHis guanylyltransferase, respectively. Hence, it is possible that these two cytosolic enzymes compete for the same tRNA. Here, we investigate substrate preferences associated with CCA and G-1-addition to yeast cytosolic tRNAHis, which might result in a temporal order to these important processing events. We show that tRNA nucleotidyltransferase accepts tRNAHis transcripts independent of the presence of G-1; however, tRNAHis guanylyltransferase clearly prefers a substrate carrying a CCA terminus. Although many tRNA maturation steps can occur in a rather random order, our data demonstrate a likely pathway where CCA-addition precedes G-1 incorporation in S. cerevisiae. Evidently, the 3'-CCA triplet and a discriminator position A73 act as positive elements for G-1 incorporation, ensuring the fidelity of G-1 addition.
Keywords: CCA-addition; G-1 residue; tRNA maturation; tRNA nucleotidyltransferase; tRNAHis; tRNAHis guanylyltransferase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
The 3' end CCA of mature tRNA is an antideterminant for eukaryotic 3'-tRNase.RNA. 1999 Feb;5(2):245-56. doi: 10.1017/s1355838299981256. RNA. 1999. PMID: 10024176 Free PMC article.
-
Life without post-transcriptional addition of G-1: two alternatives for tRNAHis identity in Eukarya.RNA. 2015 Feb;21(2):243-53. doi: 10.1261/rna.048389.114. Epub 2014 Dec 12. RNA. 2015. PMID: 25505023 Free PMC article.
-
Naturally occurring dual recognition of tRNAHis substrates with and without a universal identity element.RNA Biol. 2019 Sep;16(9):1275-1285. doi: 10.1080/15476286.2019.1626663. Epub 2019 Jun 16. RNA Biol. 2019. PMID: 31179821 Free PMC article.
-
Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review.
-
The CCA-adding enzyme: A central scrutinizer in tRNA quality control.Bioessays. 2015 Sep;37(9):975-82. doi: 10.1002/bies.201500043. Epub 2015 Jul 14. Bioessays. 2015. PMID: 26172425 Review.
Cited by
-
The life and times of a tRNA.RNA. 2023 Jul;29(7):898-957. doi: 10.1261/rna.079620.123. Epub 2023 Apr 13. RNA. 2023. PMID: 37055150 Free PMC article. Review.
-
Synthetic riboswitches for the analysis of tRNA processing by eukaryotic RNase P enzymes.RNA. 2022 Apr;28(4):551-567. doi: 10.1261/rna.078814.121. Epub 2022 Jan 12. RNA. 2022. PMID: 35022261 Free PMC article.
-
Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes.Int J Mol Sci. 2020 Jan 10;21(2):462. doi: 10.3390/ijms21020462. Int J Mol Sci. 2020. PMID: 31936900 Free PMC article.
References
-
- Hartmann R.K., Gößringer M., Späth B., Fischer S., Marchfelder A. Chapter 8 The Making of tRNAs and More—RNase P and tRNase Z. In: Condon C., editor. Molecular Biology of RNA Processing and Decay in Prokaryotes. Volume 85. Elsevier; Amsterdam, The Netherlands: 2009. pp. 319–368. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

