Saccharibacteria (TM7) in the Human Oral Microbiome
- PMID: 30894042
- PMCID: PMC6481004
- DOI: 10.1177/0022034519831671
Saccharibacteria (TM7) in the Human Oral Microbiome
Abstract
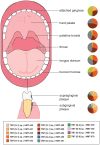
Bacteria from the Saccharibacteria phylum (formerly known as TM7) are ubiquitous members of the human oral microbiome and are part of the Candidate Phyla Radiation. Recent studies have revealed remarkable 16S rRNA diversity in environmental and mammalian host-associated members across this phylum, and their association with oral mucosal infectious diseases has been reported. However, due to their recalcitrance to conventional cultivation, TM7's physiology, lifestyle, and role in health and diseases remain elusive. The recent cultivation and characterization of Nanosynbacter lyticus type strain TM7x (HMT_952)-the first Saccharibacteria strain coisolated as an ultrasmall obligate parasite with its bacterial host from the human oral cavity-provide a rare glimpse into the novel symbiotic lifestyle of these enigmatic human-associated bacteria. TM7x is unique among all bacteria: it has an ultrasmall size and lives on the surface of its host bacterium. With a highly reduced genome, it lacks the ability to synthesize any of its own amino acids, vitamins, or cell wall precursors and must parasitize other oral bacteria. TM7x displays a highly dynamic interaction with its bacterial hosts, as reflected by the reciprocal morphologic and physiologic changes in both partners. Furthermore, depending on environmental conditions, TM7x can exhibit virulent killing of its host bacterium. Thus, Saccharibacteria potentially affect oral microbial ecology by modulating the oral microbiome structure hierarchy and functionality through affecting the bacterial host's physiology, inhibiting the host's growth dynamics, or affecting the relative abundance of the host via direct killing. At this time, several other uncharacterized members of this phylum have been detected in various human body sites at high prevalence. In the oral cavity alone, at least 6 distinct groups vary widely in relative abundance across anatomic sites. Here, we review the current knowledge on the diversity and unique biology of this recently uncovered group of ultrasmall bacteria.
Keywords: candidate phyla radiation; epiparasite; interspecies interaction; periodontitis; symbiont; ultra-small bacteria.
Figures


References
-
- Abrams M, Barton DE, Vandaei E, Romero D, Caldwell A, Ouverney CC. 2012. Genomic characteristics of an environmental microbial community harboring a novel human uncultivated TM7 bacterium associated with oral diseases. Open Access Sci Rep. 1:276.
-
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 31(6):533–538. - PubMed
-
- Attar N. 2016. Bacterial evolution: CPR breathes new air into the tree of life. Nat Rev Microbiol. 14(6):332–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

