KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy
- PMID: 30894058
- PMCID: PMC6844519
- DOI: 10.1080/15548627.2019.1596491
KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy
Abstract
Macroautophagy/autophagy is involved in myeloid cellular repair, destruction, and osteoclast differentiation; conversely, KLF2 (kruppel-like factor 2 [lung]) regulates myeloid cell activation and differentiation. To investigate the specific role of KLF2 in autophagy, osteoclastic differentiation was induced in monocytes in presence or absence of the autophagy inhibitor 3-methyladenine (3-MA), KLF2 inducer geranylgeranyl transferase inhibitor (GGTI298), and adenoviral overexpression of KLF2. We found that the number of autophagic cells and multinucleated osteoclasts were significantly decreased in presence of 3-MA, GGTI298, and KLF2 overexpressed cells indicating involvement of KLF2 in these processes. In addition, autophagy-related protein molecules were significantly decreased after induction of KLF2 during the course of osteoclastic differentiation. Furthermore, induction of arthritis in mice reduced the level of Klf2 in monocytes, and enhanced autophagy during osteoclastic differentiation. Mechanistically, knocking down of KLF2 increased the level of Beclin1 (BECN1) expression, and conversely, KLF2 over-expression reduced the level of BECN1 in monocytes. Moreover, 3-MA and GGTI298 both reduced myeloid cell proliferation concomitantly upregulating senescence-related molecules (CDKN1A/p21 and CDKN1B/p27kip1). We further confirmed epigenetic regulation of Becn1 by modulating Klf2; knocking down of Klf2 increased the levels of histone activation marks H3K9 and H4K8 acetylation in the promoter region of Becn1; and overexpression of Klf2 decreased the levels of H4K8 and H3K9 acetylation. In addition, osteoclastic differentiation also increased levels of H3K9 and H4K8 acetylation in the promoter region of Becn1. Together these findings for the first time revealed that Klf2 critically regulates Becn1-mediated autophagy process during osteoclastogenesis.Abbreviations: ACP5/TRAP: acid phosphatase 5, tartrate resistant; Ad-KLF2: adenoviral construct of KLF2; ATG3: autophagy related 3; ATG5: autophagy related 5; ATG7: autophagy related 7; ATG12: autophagy related 12; BECN1: beclin 1, autophagy related; C57BL/6: inbred mouse strain C57 black 6; ChIP: chromatin immunoprecipitation; CSF1/MCSF: colony stimulating factor 1 (macrophage); CTSK: cathepsin K; EV: empty vector; GGTI298: geranylgeranyl transferase inhibitor; H3K9Ac: histone H3 lysine 9 acetylation; H4K8Ac: histone H4 lysine 8 acetylation; K/BxN mice: T cell receptor (TCR) transgene KRN and the MHC class II molecule A(g7) generates K/BxN mice; KLF2: kruppel-like factor 2 (lung); 3MA: 3-methyladenine; MAP1LC3B/LC3B: microtubule-associated protein 1 light chain 3 beta; MDC: monodansylcadaverine; NFATc1: nuclear factor of activated T cells 1; NFKB: nuclear factor of kappa light polypeptide gene enhancer in B cells; p21/CDKN1A: cyclin dependent kinase inhibitor 1A; p27kip1/CDKN1B: cyclin-dependent kinase inhibitor 1B; PCR: polymerase chain reaction; PtdIns3K: phosphoinositide 3-kinase; RA: rheumatoid arthritis; siKlf2: small interfering KLF2 ribonucleic acid; NS: non-specific; RAW 264.7: abelson murine leukemia virus transformed macrophage cell line; TNFSF11/RANKL: tumor necrosis factor (ligand) superfamily, member 11; TSS: transcriptional start site; UCSC: University of California, Santa Cruz.
Keywords: Autophagy; KLF2; histone acetylation; monocytes; osteoclast differentiation.
Figures




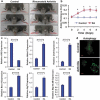


Similar articles
-
Induction of Krüppel-like factor 2 reduces K/BxN serum-induced arthritis.J Cell Mol Med. 2019 Feb;23(2):1386-1395. doi: 10.1111/jcmm.14041. Epub 2018 Dec 3. J Cell Mol Med. 2019. PMID: 30506878 Free PMC article.
-
Puerarin inhibits the osteoclastogenesis by inhibiting RANKL-dependent and -independent autophagic responses.BMC Complement Altern Med. 2019 Oct 15;19(1):269. doi: 10.1186/s12906-019-2691-5. BMC Complement Altern Med. 2019. PMID: 31615565 Free PMC article.
-
Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis.Autophagy. 2020 Jun;16(6):1021-1043. doi: 10.1080/15548627.2019.1658436. Epub 2019 Sep 8. Autophagy. 2020. PMID: 31462144 Free PMC article.
-
Transcriptional Regulation of Osteoclastogenesis: The Emerging Role of KLF2.Front Immunol. 2020 May 13;11:937. doi: 10.3389/fimmu.2020.00937. eCollection 2020. Front Immunol. 2020. PMID: 32477372 Free PMC article. Review.
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
Cited by
-
EZH2 regulates the balance between osteoclast and osteoblast differentiation to inhibit arthritis-induced bone destruction.Genes Immun. 2022 Jun;23(3-4):141-148. doi: 10.1038/s41435-022-00174-8. Epub 2022 May 17. Genes Immun. 2022. PMID: 35581496
-
Autophagy Modulation as a Potential Therapeutic Strategy in Osteosarcoma: Current Insights and Future Perspectives.Int J Mol Sci. 2023 Sep 7;24(18):13827. doi: 10.3390/ijms241813827. Int J Mol Sci. 2023. PMID: 37762129 Free PMC article. Review.
-
KLF2 Regulates Neural Differentiation of Dental Pulp-derived Stem Cells by Modulating Autophagy and Mitophagy.Stem Cell Rev Rep. 2023 Nov;19(8):2886-2900. doi: 10.1007/s12015-023-10607-0. Epub 2023 Aug 29. Stem Cell Rev Rep. 2023. PMID: 37642902
-
Identification of Ferroptosis-Related Biomarkers for Diagnosis and Molecular Classification of Staphylococcus aureus-Induced Osteomyelitis.J Inflamm Res. 2023 Apr 26;16:1805-1823. doi: 10.2147/JIR.S406562. eCollection 2023. J Inflamm Res. 2023. PMID: 37131411 Free PMC article.
-
Hepatocyte growth factor overexpression promotes osteoclastogenesis and exacerbates bone loss in CIA mice.J Orthop Translat. 2020 Dec 10;27:9-16. doi: 10.1016/j.jot.2020.10.011. eCollection 2021 Mar. J Orthop Translat. 2020. PMID: 33344167 Free PMC article.
References
-
- Imboden JB. The immunopathogenesis of rheumatoid arthritis. Annu Rev Pathol. 2009;4:417–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
