Universal human papillomavirus typing by whole genome sequencing following target enrichment: evaluation of assay reproducibility and limit of detection
- PMID: 30894118
- PMCID: PMC6425667
- DOI: 10.1186/s12864-019-5598-0
Universal human papillomavirus typing by whole genome sequencing following target enrichment: evaluation of assay reproducibility and limit of detection
Abstract
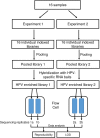
Background: We recently described a method for unbiased detection of all known human papillomaviruses (HPV) types with the potential for the determination of their variant and integration from the resulting whole genome sequence data. Considering the complex workflow for target-enriched next generation sequencing (NGS), we focused on the reproducibility and limit of detection (LOD) of this new universal HPV typing assay in this study.
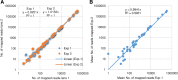
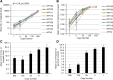
Results: We evaluated the reproducibility and LOD for HPV genotyping based on our recently published method that used RNA-baits targeting whole genomes of 191 HPV types, Agilent SureSelect protocol for target enrichment and Illumina HiSeq 2500 for sequencing (eWGS, enriched whole genome sequencing). Two libraries, prepared from pooled plasmids representing 9 vaccine HPV types at varying input (1-625 copies/reaction), were sequenced twice giving four replicates for evaluating reproducibility and LOD. eWGS showed high correlation in the number of reads mapped to HPV reference genomes between the two flow-cell lanes within (R2 = 1) and between experiments (R2 = 0.99). The number of mapped reads was positively correlated to copy number (β = 13.9, p < 0.0001). The limit of blank (LOB) could be calculated based on mapped reads to HPV types not included in each sample. HPV genotyping was reproducible for all 9 types at 625 copies using multiple cut-off criteria but LOD was 25 copies based on number of reads above LOB even when multiple types were present. eWGS showed no bias for HPV genotyping under single or multiple infection (p = 0.16-0.99).
Conclusions: The universal eWGS method for HPV genotyping has sensitivity, competitive with widely used consensus PCR methods with reduced type competition, and with the potential for determination of variant and integration status. The protocol used in this study, using defined samples varying in complexity and copy number, analyzed in replicate and duplicate assays, is applicable to most WGS methods.
Keywords: HPV typing; LOD; NGS; Reproducibility; Target enrichment.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Bioinformatics Pipeline for Human Papillomavirus Short Read Genomic Sequences Classification Using Support Vector Machine.Viruses. 2020 Jun 30;12(7):710. doi: 10.3390/v12070710. Viruses. 2020. PMID: 32629900 Free PMC article.
-
Universal Human Papillomavirus Typing Assay: Whole-Genome Sequencing following Target Enrichment.J Clin Microbiol. 2017 Mar;55(3):811-823. doi: 10.1128/JCM.02132-16. Epub 2016 Dec 14. J Clin Microbiol. 2017. PMID: 27974548 Free PMC article.
-
Next generation sequencing for human papillomavirus genotyping.J Clin Virol. 2013 Oct;58(2):437-42. doi: 10.1016/j.jcv.2013.07.013. Epub 2013 Aug 8. J Clin Virol. 2013. PMID: 23932809
-
Human papillomavirus genotyping by 454 next generation sequencing technology.J Clin Virol. 2011 Oct;52(2):93-7. doi: 10.1016/j.jcv.2011.07.006. Epub 2011 Jul 29. J Clin Virol. 2011. PMID: 21802982
-
Molecular methods for identification and characterization of novel papillomaviruses.Clin Microbiol Infect. 2015 Sep;21(9):808-16. doi: 10.1016/j.cmi.2015.05.011. Epub 2015 May 21. Clin Microbiol Infect. 2015. PMID: 26003284 Review.
Cited by
-
Human Papillomavirus Detection by Whole-Genome Next-Generation Sequencing: Importance of Validation and Quality Assurance Procedures.Viruses. 2021 Jul 8;13(7):1323. doi: 10.3390/v13071323. Viruses. 2021. PMID: 34372528 Free PMC article.
-
Hybrid-Capture Target Enrichment in Human Pathogens: Identification, Evolution, Biosurveillance, and Genomic Epidemiology.Pathogens. 2024 Mar 23;13(4):275. doi: 10.3390/pathogens13040275. Pathogens. 2024. PMID: 38668230 Free PMC article. Review.
-
Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing.Viruses. 2020 Dec 14;12(12):1437. doi: 10.3390/v12121437. Viruses. 2020. PMID: 33327447 Free PMC article.
-
Bioinformatics Pipeline for Human Papillomavirus Short Read Genomic Sequences Classification Using Support Vector Machine.Viruses. 2020 Jun 30;12(7):710. doi: 10.3390/v12070710. Viruses. 2020. PMID: 32629900 Free PMC article.
-
Nested PCR followed by NGS: Validation and application for HPV genotyping of Tunisian cervical samples.PLoS One. 2021 Aug 11;16(8):e0255914. doi: 10.1371/journal.pone.0255914. eCollection 2021. PLoS One. 2021. PMID: 34379683 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

