Designing a rigorous microscopy experiment: Validating methods and avoiding bias
- PMID: 30894402
- PMCID: PMC6504886
- DOI: 10.1083/jcb.201812109
Designing a rigorous microscopy experiment: Validating methods and avoiding bias
Abstract
Images generated by a microscope are never a perfect representation of the biological specimen. Microscopes and specimen preparation methods are prone to error and can impart images with unintended attributes that might be misconstrued as belonging to the biological specimen. In addition, our brains are wired to quickly interpret what we see, and with an unconscious bias toward that which makes the most sense to us based on our current understanding. Unaddressed errors in microscopy images combined with the bias we bring to visual interpretation of images can lead to false conclusions and irreproducible imaging data. Here we review important aspects of designing a rigorous light microscopy experiment: validation of methods used to prepare samples and of imaging system performance, identification and correction of errors, and strategies for avoiding bias in the acquisition and analysis of images.
© 2019 Jost and Waters.
Figures
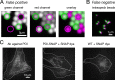
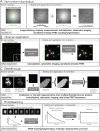
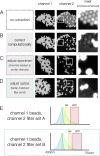


References
-
- Allan V.J., editor. 2000. Protein localization by fluorescent microscopy : a practical approach. Oxford University Press, Oxford.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

