Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
- PMID: 30894413
- PMCID: PMC6509485
- DOI: 10.1074/jbc.RA119.007847
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
Abstract
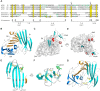
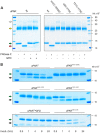
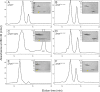
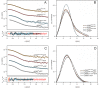
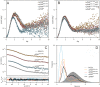
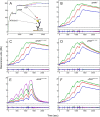
The urokinase receptor (uPAR) is a founding member of a small protein family with multiple Ly6/uPAR (LU) domains. The motif defining these LU domains contains five plesiotypic disulfide bonds stabilizing its prototypical three-fingered fold having three protruding loops. Notwithstanding the detailed knowledge on structure-function relationships in uPAR, one puzzling enigma remains unexplored. Why does the first LU domain in uPAR (DI) lack one of its consensus disulfide bonds, when the absence of this particular disulfide bond impairs the correct folding of other single LU domain-containing proteins? Here, using a variety of contemporary biophysical methods, we found that reintroducing the two missing half-cystines in uPAR DI caused the spontaneous formation of the corresponding consensus 7-8 LU domain disulfide bond. Importantly, constraints due to this cross-link impaired (i) the binding of uPAR to its primary ligand urokinase and (ii) the flexible interdomain assembly of the three LU domains in uPAR. We conclude that the evolutionary deletion of this particular disulfide bond in uPAR DI may have enabled the assembly of a high-affinity urokinase-binding cavity involving all three LU domains in uPAR. Of note, an analogous neofunctionalization occurred in snake venom α-neurotoxins upon loss of another pair of the plesiotypic LU domain half-cystines. In summary, elimination of the 7-8 consensus disulfide bond in the first LU domain of uPAR did have significant functional and structural consequences.
Keywords: LU domain; disulfide; fibrinolysis; hydrogen exchange mass spectrometry; plasminogen regulation; protein evolution; receptor structure-function; small-angle X-ray scattering (SAXS); surface plasmon resonance (SPR); urokinase receptor.
© 2019 Leth et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Ploug M., Rønne E., Behrendt N., Jensen A. L., Blasi F., and Danø K. (1991) Cellular receptor for urokinase plasminogen activator: carboxyl-terminal processing and membrane anchoring by glycosylphosphatidylinositol. J. Biol. Chem. 266, 1926–1933 - PubMed
-
- Kjaergaard M., Hansen L. V., Jacobsen B., Gardsvoll H., and Ploug M. (2008) Structure and ligand interactions of the urokinase receptor (uPAR). Front. Biosci. 13, 5441–5461 - PubMed
-
- Connolly B. M., Choi E. Y., Gårdsvoll H., Bey A. L., Currie B. M., Chavakis T., Liu S., Molinolo A., Ploug M., Leppla S. H., and Bugge T. H. (2010) Selective abrogation of the uPA-uPAR interaction in vivo reveals a novel role in suppression of fibrin-associated inflammation. Blood 116, 1593–1603 10.1182/blood-2010-03-276642 - DOI - PMC - PubMed
-
- Gårdsvoll H., Jacobsen B., Kriegbaum M. C., Behrendt N., Engelholm L., Østergaard S., and Ploug M. (2011) Conformational regulation of urokinase receptor function: impact of receptor occupancy and epitope-mapped monoclonal antibodies on lamellipodia induction. J. Biol. Chem. 286, 33544–33556 10.1074/jbc.M111.220087 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

