Mutations That Increase the Stability of the Postfusion gp41 Conformation of the HIV-1 Envelope Glycoprotein Are Selected by both an X4 and R5 HIV-1 Virus To Escape Fusion Inhibitors Corresponding to Heptad Repeat 1 of gp41, but the gp120 Adaptive Mutations Differ between the Two Viruses
- PMID: 30894471
- PMCID: PMC6532104
- DOI: 10.1128/JVI.00142-19
Mutations That Increase the Stability of the Postfusion gp41 Conformation of the HIV-1 Envelope Glycoprotein Are Selected by both an X4 and R5 HIV-1 Virus To Escape Fusion Inhibitors Corresponding to Heptad Repeat 1 of gp41, but the gp120 Adaptive Mutations Differ between the Two Viruses
Abstract
Binding of the gp120 surface subunit of the envelope glycoprotein (Env) of HIV-1 to CD4 and chemokine receptors on target cells triggers refolding of the gp41 transmembrane subunit into a six-helix bundle (6HB) that promotes fusion between virus and host cell membranes. To elucidate details of Env entry and potential differences between viruses that use CXCR4 (X4) or CCR5 (R5) coreceptors, we generated viruses that are resistant to peptide fusion inhibitors corresponding to the first heptad repeat region (HR1) of gp41 that target fusion-intermediate conformations of Env. Previously we reported that an R5 virus selected two resistance pathways, each defined by an early gp41 resistance mutation in either HR1 or the second heptad repeat (HR2), to escape inhibition by an HR1 peptide, but preferentially selected the HR1 pathway to escape inhibition by a trimer-stabilized HR1 peptide. Here, we report that an X4 virus selected the same HR1 and HR2 resistance pathways as the R5 virus to escape inhibition by the HR1 peptide. However, in contrast to the R5 virus, the X4 virus selected a unique mutation in HR2 to escape inhibition by the trimer-stabilized peptide. Significantly, both of these X4 and R5 viruses acquired gp41 resistance mutations that improved the thermostability of the six-helix bundle, but they selected different gp120 adaptive mutations. These findings show that these X4 and R5 viruses use a similar resistance mechanism to escape from HR1 peptide inhibition but different gp120-gp41 interactions to regulate Env conformational changes.IMPORTANCE HIV-1 fuses with cells when the gp41 subunit of Env refolds into a 6HB after binding to cellular receptors. Peptides corresponding to HR1 or HR2 interrupt gp41 refolding and inhibit HIV infection. Previously, we found that a CCR5 coreceptor-tropic HIV-1 acquired a key HR1 or HR2 resistance mutation to escape HR1 peptide inhibitors but only the key HR1 mutation to escape a trimer-stabilized HR1 peptide inhibitor. Here, we report that a CXCR4 coreceptor-tropic HIV-1 selected the same key HR1 or HR2 mutations to escape inhibition by the HR1 peptide but different combinations of HR1 and HR2 mutations to escape the trimer-stabilized HR1 peptide. All gp41 mutations enhance 6HB stability to outcompete inhibitors, but gp120 adaptive mutations differed between these R5 and X4 viruses, providing new insights into gp120-gp41 functional interactions affecting Env refolding during HIV entry.
Keywords: HIV-1; conformational changes; fusion; fusion inhibitor; gp41; resistance.
Copyright © 2019 American Society for Microbiology.
Figures
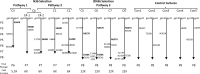
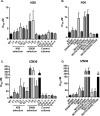




Similar articles
-
Mutations of Glu560 within HIV-1 Envelope Glycoprotein N-terminal heptad repeat region contribute to resistance to peptide inhibitors of virus entry.Retrovirology. 2019 Dec 3;16(1):36. doi: 10.1186/s12977-019-0496-8. Retrovirology. 2019. PMID: 31796053 Free PMC article.
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
Trimeric, coiled-coil extension on peptide fusion inhibitor of HIV-1 influences selection of resistance pathways.J Biol Chem. 2012 Mar 9;287(11):8297-309. doi: 10.1074/jbc.M111.324483. Epub 2012 Jan 10. J Biol Chem. 2012. PMID: 22235115 Free PMC article.
-
HIV-1 gp41 fusion intermediate: a target for HIV therapeutics.J Formos Med Assoc. 2010 Feb;109(2):94-105. doi: 10.1016/S0929-6646(10)60029-0. J Formos Med Assoc. 2010. PMID: 20206833 Review.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
Cited by
-
Heptad repeat 1-derived N peptide inhibitors improve broad-spectrum anti-HIV-1 activity.Curr Res Microb Sci. 2025 Feb 21;8:100364. doi: 10.1016/j.crmicr.2025.100364. eCollection 2025. Curr Res Microb Sci. 2025. PMID: 40093556 Free PMC article.
-
Mutations of Glu560 within HIV-1 Envelope Glycoprotein N-terminal heptad repeat region contribute to resistance to peptide inhibitors of virus entry.Retrovirology. 2019 Dec 3;16(1):36. doi: 10.1186/s12977-019-0496-8. Retrovirology. 2019. PMID: 31796053 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

