An Immunodominant and Conserved B-Cell Epitope in the Envelope of Simian Foamy Virus Recognized by Humans Infected with Zoonotic Strains from Apes
- PMID: 30894477
- PMCID: PMC6532072
- DOI: 10.1128/JVI.00068-19
An Immunodominant and Conserved B-Cell Epitope in the Envelope of Simian Foamy Virus Recognized by Humans Infected with Zoonotic Strains from Apes
Abstract
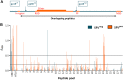
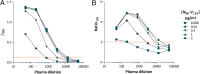
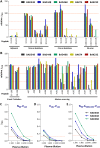
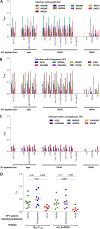
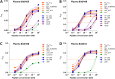
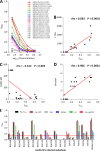
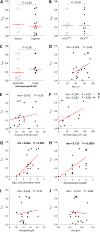
Cross-species transmission of simian foamy viruses (SFVs) from nonhuman primates (NHPs) to humans is currently ongoing. These zoonotic retroviruses establish lifelong persistent infection in their human hosts. SFV are apparently nonpathogenic in vivo, with ubiquitous in vitro tropism. Here, we aimed to identify envelope B-cell epitopes that are recognized following a zoonotic SFV infection. We screened a library of 169 peptides covering the external portion of the envelope from the prototype foamy virus (SFVpsc_huHSRV.13) for recognition by samples from 52 Central African hunters (16 uninfected and 36 infected with chimpanzee, gorilla, or Cercopithecus SFV). We demonstrate the specific recognition of peptide N96-V110 located in the leader peptide, gp18LP Forty-three variant peptides with truncations, alanine substitutions, or amino acid changes found in other SFV species were tested. We mapped the epitope between positions 98 and 108 and defined six amino acids essential for recognition. Most plasma samples from SFV-infected humans cross-reacted with sequences from apes and Old World monkey SFV species. The magnitude of binding to peptide N96-V110 was significantly higher for samples of individuals infected with a chimpanzee or gorilla SFV than those infected with a Cercopithecus SFV. In conclusion, we have been the first to define an immunodominant B-cell epitope recognized by humans following zoonotic SFV infection.IMPORTANCE Foamy viruses are the oldest known retroviruses and have been mostly described to be nonpathogenic in their natural animal hosts. SFVs can be transmitted to humans, in whom they establish persistent infection, like the simian lenti- and deltaviruses that led to the emergence of two major human pathogens, human immunodeficiency virus type 1 and human T-lymphotropic virus type 1. This is the first identification of an SFV-specific B-cell epitope recognized by human plasma samples. The immunodominant epitope lies in gp18LP, probably at the base of the envelope trimers. The NHP species the most genetically related to humans transmitted SFV strains that induced the strongest antibody responses. Importantly, this epitope is well conserved across SFV species that infect African and Asian NHPs.
Keywords: antibody; emergence; retrovirus; zoonotic infections.
Copyright © 2019 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

