Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms
- PMID: 30894479
- PMCID: PMC6452659
- DOI: 10.1073/pnas.1818486116
Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms
Abstract
It is often stated that polymorphisms for mutations affecting fitness of males and females in opposite directions [sexually antagonistic (SA) polymorphisms] are the main selective force for the evolution of recombination suppression between sex chromosomes. However, empirical evidence to discriminate between different hypotheses is difficult to obtain. We report genetic mapping results in laboratory-raised families of the guppy (Poecilia reticulata), a sexually dimorphic fish with SA polymorphisms for male coloration genes, mostly on the sex chromosomes. Comparison of the genetic and physical maps shows that crossovers are distributed very differently in the two sexes (heterochiasmy); in male meiosis, they are restricted to the termini of all four chromosomes studied, including chromosome 12, which carries the sex-determining locus. Genome resequencing of male and female guppies from a population also indicates sex linkage of variants across almost the entire chromosome 12. More than 90% of the chromosome carrying the male-determining locus is therefore transmitted largely through the male lineage. A lack of heterochiasmy in a related fish species suggests that it originated recently in the lineage leading to the guppy. Our findings do not support the hypothesis that suppressed recombination evolved in response to the presence of SA polymorphisms. Instead, a low frequency of recombination on a chromosome that carries a male-determining locus and has not undergone genetic degeneration has probably facilitated the establishment of male-beneficial coloration polymorphisms.
Keywords: crossing over; genetic maps; guppies; sex chromosomes; sexual antagonism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
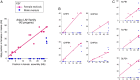
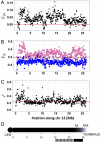

Comment in
-
Reply to Wright et al.: How to explain the absence of extensive Y-specific regions in the guppy sex chromosomes.Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12609-12610. doi: 10.1073/pnas.1906633116. Epub 2019 Jun 18. Proc Natl Acad Sci U S A. 2019. PMID: 31213530 Free PMC article. No abstract available.
-
On the power to detect rare recombination events.Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12607-12608. doi: 10.1073/pnas.1905555116. Epub 2019 Jun 18. Proc Natl Acad Sci U S A. 2019. PMID: 31213531 Free PMC article. No abstract available.
References
-
- Muller HJ. Some genetic aspects of sex. Am Nat. 1932;66:118–138.
-
- Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin/Cummings; Menlo Park, CA: 1983.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

