Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy
- PMID: 30894502
- PMCID: PMC6522349
- DOI: 10.1126/scitranslmed.aar5012
Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy
Abstract
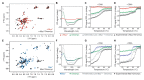
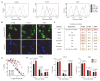
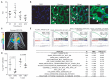
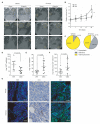
Inhibiting MYC has long been considered unfeasible, although its key role in human cancers makes it a desirable target for therapeutic intervention. One reason for its perceived undruggability was the fear of catastrophic side effects in normal tissues. However, we previously designed a dominant-negative form of MYC called Omomyc and used its conditional transgenic expression to inhibit MYC function both in vitro and in vivo. MYC inhibition by Omomyc exerted a potent therapeutic impact in various mouse models of cancer, causing only mild, well-tolerated, and reversible side effects. Nevertheless, Omomyc has been so far considered only a proof of principle. In contrast with that preconceived notion, here, we show that the purified Omomyc mini-protein itself spontaneously penetrates into cancer cells and effectively interferes with MYC transcriptional activity therein. Efficacy of the Omomyc mini-protein in various experimental models of non-small cell lung cancer harboring different oncogenic mutation profiles establishes its therapeutic potential after both direct tissue delivery and systemic administration, providing evidence that the Omomyc mini-protein is an effective MYC inhibitor worthy of clinical development.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

Comment in
-
Long path to MYC inhibition approaches clinical trials.Nat Rev Cancer. 2019 May;19(5):252. doi: 10.1038/s41568-019-0141-9. Nat Rev Cancer. 2019. PMID: 30967650 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

