Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase
- PMID: 30894746
- PMCID: PMC7611299
- DOI: 10.1038/s41586-019-1030-9
Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase
Abstract
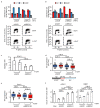
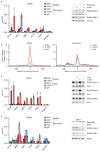
MYC is an oncogenic transcription factor that binds globally to active promoters and promotes transcriptional elongation by RNA polymerase II (RNAPII)1,2. Deregulated expression of the paralogous protein MYCN drives the development of neuronal and neuroendocrine tumours and is often associated with a particularly poor prognosis3. Here we show that, similar to MYC, activation of MYCN in human neuroblastoma cells induces escape of RNAPII from promoters. If the release of RNAPII from transcriptional pause sites (pause release) fails, MYCN recruits BRCA1 to promoter-proximal regions. Recruitment of BRCA1 prevents MYCN-dependent accumulation of stalled RNAPII and enhances transcriptional activation by MYCN. Mechanistically, BRCA1 stabilizes mRNA decapping complexes and enables MYCN to suppress R-loop formation in promoter-proximal regions. Recruitment of BRCA1 requires the ubiquitin-specific protease USP11, which binds specifically to MYCN when MYCN is dephosphorylated at Thr58. USP11, BRCA1 and MYCN stabilize each other on chromatin, preventing proteasomal turnover of MYCN. Because BRCA1 is highly expressed in neuronal progenitor cells during early development4 and MYC is less efficient than MYCN in recruiting BRCA1, our findings indicate that a cell-lineage-specific stress response enables MYCN-driven tumours to cope with deregulated RNAPII function.
Conflict of interest statement
The authors do not declare a conflict of interest.
Figures












References
-
- Rickman DS, Schulte JH, Eilers M. The Expanding World of N-MYC-Driven Tumors. Cancer Discov. 2018;8:150–163. doi: 10.1158/2159-8290.CD-17-0273. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

