The expanding landscape of 'oncohistone' mutations in human cancers
- PMID: 30894748
- PMCID: PMC6512987
- DOI: 10.1038/s41586-019-1038-1
The expanding landscape of 'oncohistone' mutations in human cancers
Abstract
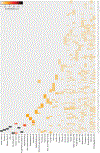
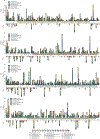
Mutations in epigenetic pathways are common oncogenic drivers. Histones, the fundamental substrates for chromatin-modifying and remodelling enzymes, are mutated in tumours including gliomas, sarcomas, head and neck cancers, and carcinosarcomas. Classical 'oncohistone' mutations occur in the N-terminal tail of histone H3 and affect the function of polycomb repressor complexes 1 and 2 (PRC1 and PRC2). However, the prevalence and function of histone mutations in other tumour contexts is unknown. Here we show that somatic histone mutations occur in approximately 4% (at a conservative estimate) of diverse tumour types and in crucial regions of histone proteins. Mutations occur in all four core histones, in both the N-terminal tails and globular histone fold domains, and at or near residues that contain important post-translational modifications. Many globular domain mutations are homologous to yeast mutants that abrogate the need for SWI/SNF function, occur in the key regulatory 'acidic patch' of histones H2A and H2B, or are predicted to disrupt the H2B-H4 interface. The histone mutation dataset and the hypotheses presented here on the effect of the mutations on important chromatin functions should serve as a resource and starting point for the chromatin and cancer biology fields in exploring an expanding role of histone mutations in cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



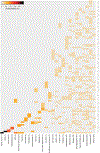








References
-
- Luger K, Mäder a W., Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). - PubMed
-
- Allis CD, Jenuwein T & Reinberg D Epigenetics Cold Spring Harb. Lab. Press. Cold Spring Harb; New York: (2007).
-
- Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45 (2000). - PubMed
-
- Jenuwein T & Allis CD Allis: Translating the histone code. Science 293, 1074–1080 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

