Neoantigen-directed immune escape in lung cancer evolution
- PMID: 30894752
- PMCID: PMC6954100
- DOI: 10.1038/s41586-019-1032-7
Neoantigen-directed immune escape in lung cancer evolution
Abstract
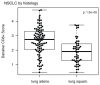
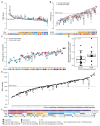
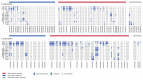
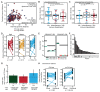
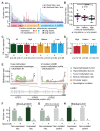
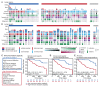
The interplay between an evolving cancer and a dynamic immune microenvironment remains unclear. Here we analyse 258 regions from 88 early-stage, untreated non-small-cell lung cancers using RNA sequencing and histopathology-assessed tumour-infiltrating lymphocyte estimates. Immune infiltration varied both between and within tumours, with different mechanisms of neoantigen presentation dysfunction enriched in distinct immune microenvironments. Sparsely infiltrated tumours exhibited a waning of neoantigen editing during tumour evolution, indicative of historical immune editing, or copy-number loss of previously clonal neoantigens. Immune-infiltrated tumour regions exhibited ongoing immunoediting, with either loss of heterozygosity in human leukocyte antigens or depletion of expressed neoantigens. We identified promoter hypermethylation of genes that contain neoantigenic mutations as an epigenetic mechanism of immunoediting. Our results suggest that the immune microenvironment exerts a strong selection pressure in early-stage, untreated non-small-cell lung cancers that produces multiple routes to immune evasion, which are clinically relevant and forecast poor disease-free survival.
Conflict of interest statement
The authors declare competing financial interests: C.S. receives grant support from Pfizer, AstraZeneca, BMS, and Ventana. C.S. has consulted for Boehringer Ingelheim, Eli Lily, Servier, Novartis, Roche-Genentech, GlaxoSmithKline, Pfizer, BMS, Celgene, AstraZeneca, Illumina, and Sarah Cannon Research Institute. C.S. is a shareholder of Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options and is co-founder of Achilles Therapeutics. S.A.Q. is a co-founder of Achilles Therapeutics. R.R., N.M., and G.A.W. have stock options and have consulted for Achilles Therapeutics.
Figures





Comment in
-
Immunoediting defines prognosis.Nat Rev Clin Oncol. 2019 May;16(5):271. doi: 10.1038/s41571-019-0205-5. Nat Rev Clin Oncol. 2019. PMID: 30926928 No abstract available.
-
Evading the immune system.Nat Med. 2019 May;25(5):710. doi: 10.1038/s41591-019-0453-z. Nat Med. 2019. PMID: 31068709 No abstract available.
-
Lung Cancer Evolution: What's Immunity Got to Do with It?Cancer Cell. 2019 May 13;35(5):711-713. doi: 10.1016/j.ccell.2019.04.009. Cancer Cell. 2019. PMID: 31085173
-
Understanding the impact of immune-mediated selection on lung cancer evolution.Br J Cancer. 2021 May;124(10):1615-1617. doi: 10.1038/s41416-020-01232-6. Epub 2021 Feb 24. Br J Cancer. 2021. PMID: 33623078 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- FC001202/WT_/Wellcome Trust/United Kingdom
- 21999/CRUK_/Cancer Research UK/United Kingdom
- 28990/CRUK_/Cancer Research UK/United Kingdom
- 20466/CRUK_/Cancer Research UK/United Kingdom
- 20265/CRUK_/Cancer Research UK/United Kingdom
- 30025/CRUK_/Cancer Research UK/United Kingdom
- A22246/CRUK_/Cancer Research UK/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- 24314/CRUK_/Cancer Research UK/United Kingdom
- 16463/CRUK_/Cancer Research UK/United Kingdom
- 22246/CRUK_/Cancer Research UK/United Kingdom
- 20764/CRUK_/Cancer Research UK/United Kingdom
- FC001169/WT_/Wellcome Trust/United Kingdom
- 20465/CRUK_/Cancer Research UK/United Kingdom
- 23896/CRUK_/Cancer Research UK/United Kingdom
- ERC_/European Research Council/International
- 24956/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_1203/1/MRC_/Medical Research Council/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
- 19278/CRUK_/Cancer Research UK/United Kingdom
- A20764/CRUK_/Cancer Research UK/United Kingdom
- 29420/CRUK_/Cancer Research UK/United Kingdom
- 29569/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

