Naive and Stem Cell Memory T Cell Subset Recovery Reveals Opposing Reconstitution Patterns in CD4 and CD8 T Cells in Chronic Graft vs. Host Disease
- PMID: 30894856
- PMCID: PMC6414429
- DOI: 10.3389/fimmu.2019.00334
Naive and Stem Cell Memory T Cell Subset Recovery Reveals Opposing Reconstitution Patterns in CD4 and CD8 T Cells in Chronic Graft vs. Host Disease
Abstract
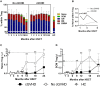
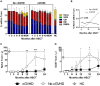
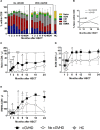
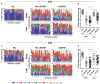
The success of allogeneic hematopoietic stem cell transplantation (allo-HSCT) in the treatment of hematological malignancies remains hampered by life-threatening chronic graft vs. host disease (cGVHD). Although multifactorial in nature, cGVHD has been associated with imbalances between effector and regulatory T cells (Treg). To further elucidate this issue, we performed a prospective analysis of patients undergoing unrelated donor allo-HSCT after a reduced intensity conditioning (RIC) regimen containing anti-thymocyte globulin (ATG) and the same GVHD prophylaxis, at a single institution. We studied T cell subset homeostasis over a 24-month follow-up after HSCT in a comparative analysis of patients with and without cGVHD. We also quantified naive and memory T cell subsets, proliferation and expression of the apoptosis-related proteins Bcl-2 and CD95. Finally, we assessed thymic function by T cell receptor excision circle (TREC) quantification and T cell receptor (TCR) diversity by TCRVβ spectratyping. While the total number of conventional CD4 (Tcon) and CD8 T cells was similar between patient groups, Treg were decreased in cGVHD patients. Interestingly, we also observed divergent patterns of Naive and Stem Cell Memory (SCM) subset recovery in Treg and Tcon compared to CD8. Patients with cGVHD showed impaired recovery of Naive and SCM Tcon and Treg, but significantly increased frequencies and absolute numbers of Naive and SCM were observed in the CD8 pool. Markedly increased EMRA CD8 T cells were also noted in cGVHD. Taken together, these results suggest that Naive, SCM and EMRA CD8 play a role in the emergence of cGHVD. Reduced Naive and recent thymic emigrant Tcon and Treg in cGVHD was likely due to impaired thymic output, as it was accompanied by decreased CD4 TREC and TCR diversity. On the other hand, CD8 TCR diversity was similar between patient groups. Furthermore, no correlation was observed between CD8 TREC content and Naive CD8 numbers, suggesting limited thymic production of Naive CD8 T cells in patients after transplant, especially in those developing cGVHD. The mechanisms behind the opposing patterns of CD4 and CD8 subset cell recovery in cGVHD remain elusive, but may be linked to thymic damage associated with the conditioning regimen and/or acute GVHD.
Keywords: Naive T cell; T lymphocyte; chronic graft vs. host disease; hematopoietic stem cell transplantation; immune reconstitution; stem cell memory.
Figures



References
-
- Passweg JR, Baldomero H, Bader P, Basak GW, Bonini C, Duarte R, et al. Is the use of unrelated donor transplantation leveling off in Europe? The 2016 European Society for Blood and Marrow Transplant activity survey report. Bone Marrow Transplant. (2018) 53:1139–48. 10.1038/s41409-018-0153-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

