A Soluble Immune Effector Binds Both Fungi and Bacteria via Separate Functional Domains
- PMID: 30894858
- PMCID: PMC6414549
- DOI: 10.3389/fimmu.2019.00369
A Soluble Immune Effector Binds Both Fungi and Bacteria via Separate Functional Domains
Abstract
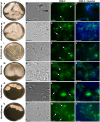
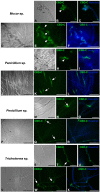
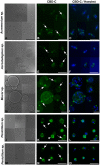
The gut microbiome of animals consists of diverse microorganisms that include both prokaryotes and eukaryotes. Complex interactions occur among these inhabitants, as well as with the immune system of the host, and profoundly influence the overall health of both the host and its microbial symbionts. Despite the enormous importance for the host to regulate its gut microbiome, the extent to which animals generate immune-related molecules with the capacity to directly influence polymicrobial interactions remains unclear. The urochordate, Ciona robusta, is a model organism that has been adapted to experimental studies of host/microbiome interactions. Ciona variable-region containing chitin-binding proteins (VCBPs) are innate immune effectors, composed of immunoglobulin (Ig) variable regions and a chitin-binding domain (CBD) and are expressed in high abundance in the gut. It was previously shown that VCBP-C binds bacteria and influences both phagocytosis by granular amoebocytes and biofilm formation via its Ig domains. We show here that the CBD of VCBP-C independently recognizes chitin molecules present in the cell walls, sporangia (spore-forming bodies), and spores of a diverse set of filamentous fungi isolated from the gut of Ciona. To our knowledge, this is the first description of a secreted Ig-containing immune molecule with the capacity to directly promote transkingdom interactions through simultaneous binding by independent structural domains and could have broad implications in modulating the establishment, succession, and homeostasis of gut microbiomes.
Keywords: Ciona; VCBP-C; fungal-immune interaction; gut immunity; host-microorganism interactions; innate immunity; mycobiota; transkingdom interactions.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

