Effect of deletion of the rgpA gene on selected virulence of Porphyromonas gingivalis
- PMID: 30894985
- PMCID: PMC6395235
- DOI: 10.1016/j.jds.2016.03.004
Effect of deletion of the rgpA gene on selected virulence of Porphyromonas gingivalis
Abstract
Background/purpose: The most potent virulence factors of the periodontal pathogen Porphyromonas gingivalis are gingipains, three cysteine proteases (RgpA, RgpB, and Kgp) that bind and cleave a wide range of host proteins. Considerable proof indicates that RgpA contributes to the entire virulence of the organism and increases the risk of periodontal disease by disrupting the host immune defense and destroying the host tissue. However, the functional significance of this proteinase is incompletely understood. It is important to analyze the effect of arginine-specific gingipain A gene (rgpA) on selected virulence and physiological properties of P. gingivalis.
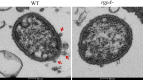
Materials and methods: Electroporation and homologous recombination were used to construct an rgpA mutant of P. gingivalis ATCC33277. The mutant was verified by polymerase chain reaction and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Cell structures of the mutant were examined by transmission electron microscopy and homotypic biofilm formation was examined by confocal laser scanning microscopy.
Results: Gene analysis revealed that the rgpA gene was deleted and replaced by a drug resistance gene marker. The defect of the gene resulted in a complete loss of RgpA proteinase, a reduction of out membrane vesicles and hemagglutination, and an increase in homotypic biofilm formation.
Conclusion: Our data indicate that an rgpA gene deficient strain of P. gingivalis is successfully isolated. RgpA may have a variety of physiological and pathological roles in P. gingivalis.
Keywords: Porphyromonas gingivalis; biofilm formation; mutant; outer membrane vesicles; rgpA.
Figures





References
-
- Slots J., Genco R.J. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984;63:412–421. - PubMed
-
- Shi Y., Kong W., Nakayama K. Human lactoferrin binds and removes the hemoglobin receptor protein of the periodontopathogen Porphyromonas gingivalis. J Biol Chem. 2000;275:30002–30008. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

