Involvement of PLAGL1/ZAC1 in hypocretin/orexin transcription
- PMID: 30896835
- PMCID: PMC6445593
- DOI: 10.3892/ijmm.2019.4143
Involvement of PLAGL1/ZAC1 in hypocretin/orexin transcription
Abstract
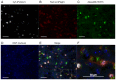
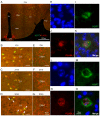
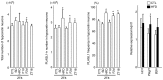
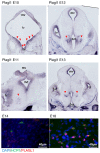
The hypocretin/orexin neuropeptide system coordinates the regulation of various physiological processes. Our previous study reported that a reduction in the expression of pleomorphic adenoma gene‑like 1 (Plagl1), which encodes a C2H2 zinc‑finger transcription factor, occurs in hypocretin neuron‑ablated transgenic mice, suggesting that PLAGL1 is co‑expressed in hypocretin neurons and regulates hypocretin transcription. The present study examined whether canonical prepro‑hypocretin transcription is functionally modulated by PLAGL1. Double immunostaining indicated that the majority of hypocretin neurons were positive for PLAGL1 immunoreactivity in the nucleus. Notably, PLAGL1 immunoreactivity in hypocretin neurons was altered in response to several conditions affecting hypocretin function. An uneven localization of PLAGL1 was detected in the nuclei of hypocretin neurons following sleep deprivation. Chromatin immunoprecipitation revealed that endogenous PLAGL1 may bind to a putative PLAGL1‑binding site in the proximal region of the hypocretin gene, in the murine hypothalamus. In addition, electroporation of the PLAGL1 expression vector into the fetal hypothalamus promoted hypothalamic hypocretin transcription. These results suggested that PLAGL1 may regulate hypothalamic hypocretin transcription.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

