Diagnostic and prognostic value of CEP55 in clear cell renal cell carcinoma as determined by bioinformatics analysis
- PMID: 30896867
- PMCID: PMC6471254
- DOI: 10.3892/mmr.2019.10042
Diagnostic and prognostic value of CEP55 in clear cell renal cell carcinoma as determined by bioinformatics analysis
Abstract
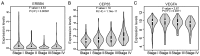
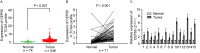
Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignant adult kidney tumor. Tumor recurrence and metastasis is the primary cause of cancer‑associated mortality in patients with ccRCC. Therefore, identification of efficient diagnostic and prognostic molecular markers may improve survival times. The GSE46699, GSE36895, GSE53000 and GSE53757 gene datasets were downloaded from the Gene Expression Omnibus database and contained 196 ccRCC samples and 164 adjacent normal kidney samples. Bioinformatics analysis was used to integrate the four microarray datasets to identify and analyze differentially expressed genes. Functional analysis revealed that there were 12 genes associated with cancer, based on the tumor‑associated gene database. Erb‑B2 receptor tyrosine kinase 4, centrosomal protein 55 (CEP55) and vascular endothelial growth factor A are oncogenes, all of which were associated with tumor stage, whereas only CEP55 was significantly associated with survival time as determined by Gene Expression Profiling Interactive Analysis. The mRNA expression levels of CEP55 in ccRCC samples were significantly higher than those observed in adjacent normal kidney tissues based on The Cancer Genome Atlas data and reverse transcription‑polymerase chain reaction results. The receiver operating characteristic curve analysis revealed that CEP55 may be considered a diagnostic biomarker for ccRCC with an area under the curve of >0.85 in the training and validation sets. High CEP55 expression was strongly associated with sex, histological grade, stage, T classification, N classification and M classification. Univariate and multivariate Cox proportional hazards analyses demonstrated that CEP55 expression was an independent risk factor for poor prognosis. In addition, gene set enrichment analysis indicated that high CEP55 expression was associated with immunization, cell adhesion, inflammation, the Janus kinase/signal transducer and activator of transcription signaling pathway and cell proliferation. In conclusion, CEP55 was increased in ccRCC samples, and may be considered a potential diagnostic and prognostic biomarker for ccRCC.
Figures





References
-
- Eikrem OS, Strauss P, Beisland C, Scherer A, Landolt L, Flatberg A, Leh S, Beisvag V, Skogstrand T, Hjelle K, et al. Development and confirmation of potential gene classifiers of human clear cell renal cell carcinoma using next-generation RNA sequencing. Scand J Urol. 2016;50:452–462. doi: 10.1080/21681805.2016.1238007. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

