Helicase/SUMO-targeted ubiquitin ligase Uls1 interacts with the Holliday junction resolvase Yen1
- PMID: 30897139
- PMCID: PMC6428284
- DOI: 10.1371/journal.pone.0214102
Helicase/SUMO-targeted ubiquitin ligase Uls1 interacts with the Holliday junction resolvase Yen1
Abstract
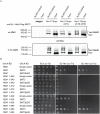
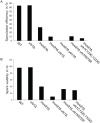
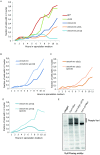
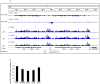
Resolution of branched DNA structures is pivotal for repair of stalled replication forks and meiotic recombination intermediates. The Yen1 nuclease cleaves both Holliday junctions and replication forks. We show that Yen1 interacts physically with Uls1, a suggested SUMO-targeted ubiquitin ligase that also contains a SWI/SNF-family ATPase-domain. Yen1 is SUMO-modified in its noncatalytic carboxyl terminus and DNA damage induces SUMOylation. SUMO-modification of Yen1 strengthens the interaction to Uls1, and mutations in SUMO interaction motifs in Uls1 weakens the interaction. However, Uls1 does not regulate the steady-state level of SUMO-modified Yen1 or chromatin-associated Yen1. In addition, SUMO-modification of Yen1 does not affect the catalytic activity in vitro. Consistent with a shared function for Uls1 and Yen1, mutations in both genes display similar phenotypes. Both uls1 and yen1 display negative genetic interactions with the alternative HJ-cleaving nuclease Mus81, manifested both in hypersensitivity to DNA damaging agents and in meiotic defects. Point mutations in ULS1 (uls1K975R and uls1C1330S, C1333S) predicted to inactivate the ATPase and ubiquitin ligase activities, respectively, are as defective as the null allele, indicating that both functions of Uls1 are essential. A micrococcal nuclease sequencing experiment showed that Uls1 had minimal effects on global nucleosome positioning/occupancy. Moreover, increased gene dosage of YEN1 partially alleviates the mus81 uls1 sensitivity to DNA damage. We suggest a preliminary model in which Uls1 acts in the same pathway as Yen1 to resolve branched DNA structures.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13(18):2339–52. Epub 1999/09/29. . - PubMed
-
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7(4):583–91. Epub 1993/04/01. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

