Computational Drug Design Applied to the Study of Metabotropic Glutamate Receptors
- PMID: 30897742
- PMCID: PMC6470756
- DOI: 10.3390/molecules24061098
Computational Drug Design Applied to the Study of Metabotropic Glutamate Receptors
Abstract
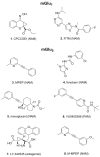
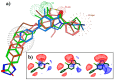
Metabotropic glutamate (mGlu) receptors are a family of eight GPCRs that are attractive drug discovery targets to modulate glutamate action and response. Here we review the application of computational methods to the study of this family of receptors. X-ray structures of the extracellular and 7-transmembrane domains have played an important role to enable structure-based modeling approaches, whilst we also discuss the successful application of ligand-based methods. We summarize the literature and highlight the areas where modeling and experiment have delivered important understanding for mGlu receptor drug discovery. Finally, we offer suggestions of future areas of opportunity for computational work.
Keywords: GPCR; allosteric modulator; homology model; ligand-based design; mGlu; mGluR; molecular dynamics; structure-based design; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

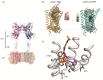
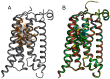

Similar articles
-
The computational modeling of allosteric modulation of metabotropic glutamate receptors.Adv Pharmacol. 2020;88:1-33. doi: 10.1016/bs.apha.2020.02.004. Epub 2020 Mar 20. Adv Pharmacol. 2020. PMID: 32416864 Review.
-
Molecular Switches of Allosteric Modulation of the Metabotropic Glutamate 2 Receptor.Structure. 2017 Jul 5;25(7):1153-1162.e4. doi: 10.1016/j.str.2017.05.021. Epub 2017 Jun 22. Structure. 2017. PMID: 28648611
-
Practical Strategies and Concepts in GPCR Allosteric Modulator Discovery: Recent Advances with Metabotropic Glutamate Receptors.Chem Rev. 2016 Jun 8;116(11):6707-41. doi: 10.1021/acs.chemrev.5b00656. Epub 2016 Feb 16. Chem Rev. 2016. PMID: 26882314 Free PMC article. Review.
-
In silico approaches towards the understanding of the structure-function relationships in metabotropic glutamate receptors (mGluRs) and other family C GPRCs.Curr Pharm Des. 2006;12(17):2159-73. doi: 10.2174/138161206777585210. Curr Pharm Des. 2006. PMID: 16796561 Review.
-
Asymmetric activation of dimeric GABAB and metabotropic glutamate receptors.Am J Physiol Cell Physiol. 2023 Jul 1;325(1):C79-C89. doi: 10.1152/ajpcell.00150.2022. Epub 2023 May 15. Am J Physiol Cell Physiol. 2023. PMID: 37184233 Review.
Cited by
-
Photoswitchable positive allosteric modulators of metabotropic glutamate receptor 4 to improve selectivity.iScience. 2024 May 28;27(6):110123. doi: 10.1016/j.isci.2024.110123. eCollection 2024 Jun 21. iScience. 2024. PMID: 38966572 Free PMC article.
-
Implications of a Neuronal Receptor Family, Metabotropic Glutamate Receptors, in Cancer Development and Progression.Cells. 2022 Sep 13;11(18):2857. doi: 10.3390/cells11182857. Cells. 2022. PMID: 36139432 Free PMC article. Review.
-
Advances in Drug Discovery and Design using Computer-aided Molecular Modeling.Curr Comput Aided Drug Des. 2024;20(5):697-710. doi: 10.2174/1573409920666230914123005. Curr Comput Aided Drug Des. 2024. PMID: 37711101 Review.
-
The role of water and protein flexibility in the structure-based virtual screening of allosteric GPCR modulators: an mGlu5 receptor case study.J Comput Aided Mol Des. 2019 Sep;33(9):787-797. doi: 10.1007/s10822-019-00224-w. Epub 2019 Sep 21. J Comput Aided Mol Des. 2019. PMID: 31542869 Free PMC article.
-
The Involvement of Neuroinflammation in the Onset and Progression of Parkinson's Disease.Int J Mol Sci. 2023 Sep 26;24(19):14582. doi: 10.3390/ijms241914582. Int J Mol Sci. 2023. PMID: 37834030 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

