Antibody repertoire analysis of mouse immunization protocols using microfluidics and molecular genomics
- PMID: 30898066
- PMCID: PMC6601537
- DOI: 10.1080/19420862.2019.1583995
Antibody repertoire analysis of mouse immunization protocols using microfluidics and molecular genomics
Abstract
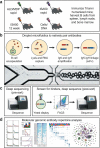
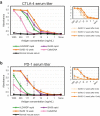
Immunization of mice followed by hybridoma or B-cell screening is one of the most common antibody discovery methods used to generate therapeutic monoclonal antibody (mAb) candidates. There are a multitude of different immunization protocols that can generate an immune response in animals. However, an extensive analysis of the antibody repertoires that these alternative immunization protocols can generate has not been performed. In this study, we immunized mice that transgenically express human antibodies with either programmed cell death 1 protein or cytotoxic T-lymphocyte associated protein 4 using four different immunization protocols, and then utilized a single cell microfluidic platform to generate tissue-specific, natively paired immunoglobulin (Ig) repertoires from each method and enriched for target-specific binders using yeast single-chain variable fragment (scFv) display. We deep sequenced the scFv repertoires from both the pre-sort and post-sort libraries. All methods and both targets yielded similar oligoclonality, variable (V) and joining (J) gene usage, and divergence from germline of enriched libraries. However, there were differences between targets and/or immunization protocols for overall clonal counts, complementarity-determining region 3 (CDR3) length, and antibody/CDR3 sequence diversity. Our data suggest that, although different immunization protocols may generate a response to an antigen, performing multiple immunization protocols in parallel can yield greater Ig diversity. We conclude that modern microfluidic methods, followed by an extensive molecular genomic analysis of antibody repertoires, can be used to quickly analyze new immunization protocols or mouse platforms.
Keywords: CTLA-4; Humanized mouse antibody repertoires; PD-1; adjuvants; deep sequencing; mouse immunization; yeast display.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
