An Allosteric Network for Spliceosome Activation Revealed by High-Throughput Suppressor Analysis in Saccharomyces cerevisiae
- PMID: 30898770
- PMCID: PMC6499515
- DOI: 10.1534/genetics.119.301922
An Allosteric Network for Spliceosome Activation Revealed by High-Throughput Suppressor Analysis in Saccharomyces cerevisiae
Abstract
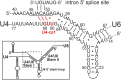
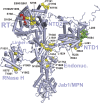
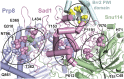
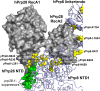
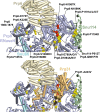
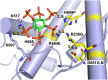
Selection of suppressor mutations that correct growth defects caused by substitutions in an RNA or protein can reveal functionally important molecular structures and interactions in living cells. This approach is particularly useful for the study of complex biological pathways involving many macromolecules, such as premessenger RNA (pre-mRNA) splicing. When a sufficiently large number of suppressor mutations is obtained and structural information is available, it is possible to generate detailed models of molecular function. However, the laborious and expensive task of identifying suppressor mutations in whole-genome selections limits the utility of this approach. Here I show that a custom targeted sequencing panel can greatly accelerate the identification of suppressor mutations in the Saccharomyces cerevisiae genome. Using a panel that targets 112 genes encoding pre-mRNA splicing factors, I identified 27 unique mutations in six protein-coding genes that each overcome the cold-sensitive block to spliceosome activation caused by a substitution in U4 small nuclear RNA. When mapped to existing structures of spliceosomal complexes, the identified suppressors implicate specific molecular contacts between the proteins Brr2, Prp6, Prp8, Prp31, Sad1, and Snu114 as functionally important in an early step of catalytic activation of the spliceosome. This approach shows great promise for elucidating the allosteric cascade of molecular interactions that direct accurate and efficient pre-mRNA splicing and should be broadly useful for understanding the dynamics of other complex biological assemblies or pathways.
Keywords: U4 snRNA; pre-mRNA splicing; spliceosome activation; suppressor selection; targeted sequencing.
Copyright © 2019 by the Genetics Society of America.
Figures


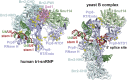
Similar articles
-
The splicing factor Prp17 interacts with the U2, U5 and U6 snRNPs and associates with the spliceosome pre- and post-catalysis.Biochem J. 2008 Dec 15;416(3):365-74. doi: 10.1042/BJ20081195. Biochem J. 2008. PMID: 18691155
-
Single molecule analysis reveals reversible and irreversible steps during spliceosome activation.Elife. 2016 May 31;5:e14166. doi: 10.7554/eLife.14166. Elife. 2016. PMID: 27244240 Free PMC article.
-
Structural dynamics of the N-terminal domain and the Switch loop of Prp8 during spliceosome assembly and activation.Nucleic Acids Res. 2018 May 4;46(8):3833-3840. doi: 10.1093/nar/gky242. Nucleic Acids Res. 2018. PMID: 29635373 Free PMC article. Review.
-
Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain.RNA. 2006 May;12(5):862-71. doi: 10.1261/rna.2319806. Epub 2006 Mar 15. RNA. 2006. PMID: 16540695 Free PMC article.
-
Mechanisms and regulation of spliceosome-mediated pre-mRNA splicing in Saccharomyces cerevisiae.Wiley Interdiscip Rev RNA. 2024 Jul-Aug;15(4):e1866. doi: 10.1002/wrna.1866. Wiley Interdiscip Rev RNA. 2024. PMID: 38972853 Free PMC article. Review.
Cited by
-
A forward genetic screen in C. elegans identifies conserved residues of spliceosomal proteins PRP8 and SNRNP200/BRR2 with a role in maintaining 5' splice site identity.Nucleic Acids Res. 2022 Nov 11;50(20):11834-11857. doi: 10.1093/nar/gkac991. Nucleic Acids Res. 2022. PMID: 36321655 Free PMC article.
-
Substitutions in RNA-binding protein Hrp1 map a potential interaction surface with the yeast RNA polymerase II elongation complex.bioRxiv [Preprint]. 2025 May 7:2025.05.07.652672. doi: 10.1101/2025.05.07.652672. bioRxiv. 2025. PMID: 40654929 Free PMC article. Preprint.
-
Control of 3' splice site selection by the yeast splicing factor Fyv6.Elife. 2024 Dec 17;13:RP100449. doi: 10.7554/eLife.100449. Elife. 2024. PMID: 39688371 Free PMC article.
-
Missplicing suppressor alleles of Arabidopsis PRE-MRNA PROCESSING FACTOR 8 increase splicing fidelity by reducing the use of novel splice sites.Nucleic Acids Res. 2022 Jun 10;50(10):5513-5527. doi: 10.1093/nar/gkac338. Nucleic Acids Res. 2022. PMID: 35639749 Free PMC article.
-
Control of 3' splice site selection by the yeast splicing factor Fyv6.bioRxiv [Preprint]. 2024 Oct 21:2024.05.04.592262. doi: 10.1101/2024.05.04.592262. bioRxiv. 2024. Update in: Elife. 2024 Dec 17;13:RP100449. doi: 10.7554/eLife.100449. PMID: 38746449 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

