Leukodystrophy-associated POLR3A mutations down-regulate the RNA polymerase III transcript and important regulatory RNA BC200
- PMID: 30898877
- PMCID: PMC6509492
- DOI: 10.1074/jbc.RA118.006271
Leukodystrophy-associated POLR3A mutations down-regulate the RNA polymerase III transcript and important regulatory RNA BC200
Abstract
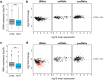
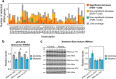
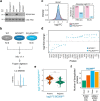
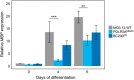
RNA polymerase III (Pol III) is an essential enzyme responsible for the synthesis of several small noncoding RNAs, a number of which are involved in mRNA translation. Recessive mutations in POLR3A, encoding the largest subunit of Pol III, cause POLR3-related hypomyelinating leukodystrophy (POLR3-HLD), characterized by deficient central nervous system myelination. Identification of the downstream effectors of pathogenic POLR3A mutations has so far been elusive. Here, we used CRISPR-Cas9 to introduce the POLR3A mutation c.2554A→G (p.M852V) into human cell lines and assessed its impact on Pol III biogenesis, nuclear import, DNA occupancy, transcription, and protein levels. Transcriptomic profiling uncovered a subset of transcripts vulnerable to Pol III hypofunction, including a global reduction in tRNA levels. The brain cytoplasmic BC200 RNA (BCYRN1), involved in translation regulation, was consistently affected in all our cellular models, including patient-derived fibroblasts. Genomic BC200 deletion in an oligodendroglial cell line led to major transcriptomic and proteomic changes, having a larger impact than those of POLR3A mutations. Upon differentiation, mRNA levels of the MBP gene, encoding myelin basic protein, were significantly decreased in POLR3A-mutant cells. Our findings provide the first evidence for impaired Pol III transcription in cellular models of POLR3-HLD and identify several candidate effectors, including BC200 RNA, having a potential role in oligodendrocyte biology and involvement in the disease.
Keywords: CRISPR/Cas; RNA polymerase III; RNA-seq; brain cytoplasmic 200 RNA (BCYRN1); leukodystrophy; myelin; oligodendrocyte; proteomics; transcription; transfer RNA (tRNA).
© 2019 Choquet et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Tetreault M., Choquet K., Orcesi S., Tonduti D., Balottin U., Teichmann M., Fribourg S., Schiffmann R., Brais B., Vanderver A., and Bernard G. (2011) Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am. J. Hum. Genet. 89, 652–655 10.1016/j.ajhg.2011.10.006 - DOI - PMC - PubMed
-
- Thiffault I., Wolf N. I., Forget D., Guerrero K., Tran L. T., Choquet K., Lavallée-Adam M., Poitras C., Brais B., Yoon G., Sztriha L., Webster R. I., Timmann D., van de Warrenburg B. P., Seeger J., et al. (2015) Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat. Commun. 6, 7623 10.1038/ncomms8623 - DOI - PMC - PubMed
-
- Bernard G., Chouery E., Putorti M. L., Tétreault M., Takanohashi A., Carosso G., Clément I., Boespflug-Tanguy O., Rodriguez D., Delague V., Abou Ghoch J., Jalkh N., Dorboz I., Fribourg S., Teichmann M., et al. (2011) Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 89, 415–423 10.1016/j.ajhg.2011.07.014 - DOI - PMC - PubMed
-
- Wolf N. I., Vanderver A., van Spaendonk R. M., Schiffmann R., Brais B., Bugiani M., Sistermans E., Catsman-Berrevoets C., Kros J. M., Pinto P. S., Pohl D., Tirupathi S., Strømme P., de Grauw T., Fribourg S., et al. (2014) Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology 83, 1898–1905 10.1212/WNL.0000000000001002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

