An Ultra-Dense Haploid Genetic Map for Evaluating the Highly Fragmented Genome Assembly of Norway Spruce (Picea abies)
- PMID: 30898899
- PMCID: PMC6505157
- DOI: 10.1534/g3.118.200840
An Ultra-Dense Haploid Genetic Map for Evaluating the Highly Fragmented Genome Assembly of Norway Spruce (Picea abies)
Abstract
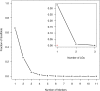
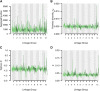
Norway spruce (Picea abies (L.) Karst.) is a conifer species of substanital economic and ecological importance. In common with most conifers, the P. abies genome is very large (∼20 Gbp) and contains a high fraction of repetitive DNA. The current P. abies genome assembly (v1.0) covers approximately 60% of the total genome size but is highly fragmented, consisting of >10 million scaffolds. The genome annotation contains 66,632 gene models that are at least partially validated (www.congenie.org), however, the fragmented nature of the assembly means that there is currently little information available on how these genes are physically distributed over the 12 P. abies chromosomes. By creating an ultra-dense genetic linkage map, we anchored and ordered scaffolds into linkage groups, which complements the fine-scale information available in assembly contigs. Our ultra-dense haploid consensus genetic map consists of 21,056 markers derived from 14,336 scaffolds that contain 17,079 gene models (25.6% of the validated gene models) that we have anchored to the 12 linkage groups. We used data from three independent component maps, as well as comparisons with previously published Picea maps to evaluate the accuracy and marker ordering of the linkage groups. We demonstrate that approximately 3.8% of the anchored scaffolds and 1.6% of the gene models covered by the consensus map have likely assembly errors as they contain genetic markers that map to different regions within or between linkage groups. We further evaluate the utility of the genetic map for the conifer research community by using an independent data set of unrelated individuals to assess genome-wide variation in genetic diversity using the genomic regions anchored to linkage groups. The results show that our map is sufficiently dense to enable detailed evolutionary analyses across the P. abies genome.
Keywords: Norway spruce; Picea abies; genetic map; genome assembly; sequence capture.
Copyright © 2019 Bernhardsson et al.
Figures




References
-
- De La Torre A. R., Birol I., Bousquet J., Ingvarsson P. K., Jansson S., et al. , 2014. Insights into Conifer Giga-Genomes. Plant Physiol. 166: 1724–1732. http://www.plantphysiol.org/content/166/4/1724.short. 10.1104/pp.114.248708 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
