Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm
- PMID: 30898902
- PMCID: PMC6445350
- DOI: 10.1126/science.aau8861
Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm
Abstract
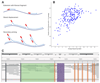
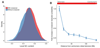
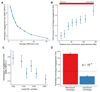
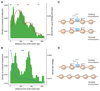
Recombination is critical to meiosis and evolution, yet many aspects of the physical exchange of DNA via crossovers remain poorly understood. We report an approach for single-cell whole-genome DNA sequencing by which we sequenced 217 individual hybrid mouse sperm, providing a kilobase-resolution genome-wide map of crossovers. Combining this map with molecular assays measuring stages of recombination, we identified factors that affect crossover probability, including PRDM9 binding on the non-initiating template homolog and telomere proximity. These factors also influence the time for sites of recombination-initiating DNA double-strand breaks to find and engage their homologs, with rapidly engaging sites more likely to form crossovers. We show that chromatin environment on the template homolog affects positioning of crossover breakpoints. Our results also offer insights into recombination in the pseudoautosomal region.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

