Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma
- PMID: 30899002
- PMCID: PMC6428878
- DOI: 10.1038/s41467-019-09179-w
Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma
Abstract
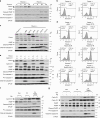
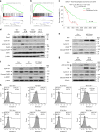
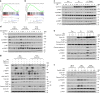
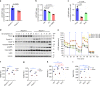
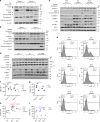
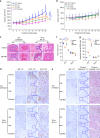
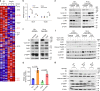
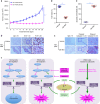
The dysregulation of Fbxo4-cyclin D1 axis occurs at high frequency in esophageal squamous cell carcinoma (ESCC), where it promotes ESCC development and progression. However, defining a therapeutic vulnerability that results from this dysregulation has remained elusive. Here we demonstrate that Rb and mTORC1 contribute to Gln-addiction upon the dysregulation of the Fbxo4-cyclin D1 axis, which leads to the reprogramming of cellular metabolism. This reprogramming is characterized by reduced energy production and increased sensitivity of ESCC cells to combined treatment with CB-839 (glutaminase 1 inhibitor) plus metformin/phenformin. Of additional importance, this combined treatment has potent efficacy in ESCC cells with acquired resistance to CDK4/6 inhibitors in vitro and in xenograft tumors. Our findings reveal a molecular basis for cancer therapy through targeting glutaminolysis and mitochondrial respiration in ESCC with dysregulated Fbxo4-cyclin D1 axis as well as cancers resistant to CDK4/6 inhibitors.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

