Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis
- PMID: 30899004
- PMCID: PMC6428843
- DOI: 10.1038/s41467-019-09222-w
Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis
Abstract
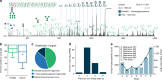
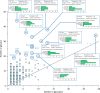
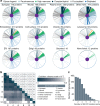
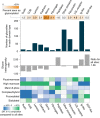
Protein glycosylation is a highly important, yet poorly understood protein post-translational modification. Thousands of possible glycan structures and compositions create potential for tremendous site heterogeneity. A lack of suitable analytical methods for large-scale analyses of intact glycopeptides has limited our abilities both to address the degree of heterogeneity across the glycoproteome and to understand how this contributes biologically to complex systems. Here we show that N-glycoproteome site-specific microheterogeneity can be captured via large-scale glycopeptide profiling methods enabled by activated ion electron transfer dissociation (AI-ETD), ultimately characterizing 1,545 N-glycosites (>5,600 unique N-glycopeptides) from mouse brain tissue. Our data reveal that N-glycosylation profiles can differ between subcellular regions and structural domains and that N-glycosite heterogeneity manifests in several different forms, including dramatic differences in glycosites on the same protein. Moreover, we use this large-scale glycoproteomic dataset to develop several visualizations that will prove useful for analyzing intact glycopeptides in future studies.
Conflict of interest statement
J.J.C. is an inventor of electron transfer dissociation and is a consultant for Thermo Fisher Scientific. The other authors declare no competing interests.
Figures



References
-
- Varki, A. et al. Essentials of Glycobiology. Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, NY, 2015).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

