The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins
- PMID: 30899024
- PMCID: PMC6428865
- DOI: 10.1038/s41467-019-09199-6
The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins
Abstract
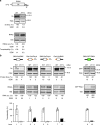
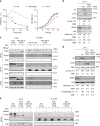
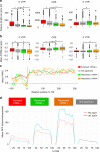
The highly conserved 5'-3' exonuclease Xrn1 regulates gene expression in eukaryotes by coupling nuclear DNA transcription to cytosolic mRNA decay. By integrating transcriptome-wide analyses of translation with biochemical and functional studies, we demonstrate an unanticipated regulatory role of Xrn1 in protein synthesis. Xrn1 promotes translation of a specific group of transcripts encoding membrane proteins. Xrn1-dependence for translation is linked to poor structural RNA contexts for translation initiation, is mediated by interactions with components of the translation initiation machinery and correlates with an Xrn1-dependence for mRNA localization at the endoplasmic reticulum, the translation compartment of membrane proteins. Importantly, for this group of mRNAs, Xrn1 stimulates transcription, mRNA translation and decay. Our results uncover a crosstalk between the three major stages of gene expression coordinated by Xrn1 to maintain appropriate levels of membrane proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

