Machine learning-powered antibiotics phenotypic drug discovery
- PMID: 30899034
- PMCID: PMC6428806
- DOI: 10.1038/s41598-019-39387-9
Machine learning-powered antibiotics phenotypic drug discovery
Abstract
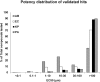
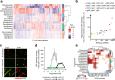
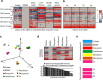
Identification of novel antibiotics remains a major challenge for drug discovery. The present study explores use of phenotypic readouts beyond classical antibacterial growth inhibition adopting a combined multiparametric high content screening and genomic approach. Deployment of the semi-automated bacterial phenotypic fingerprint (BPF) profiling platform in conjunction with a machine learning-powered dataset analysis, effectively allowed us to narrow down, compare and predict compound mode of action (MoA). The method identifies weak antibacterial hits allowing full exploitation of low potency hits frequently discovered by routine antibacterial screening. We demonstrate that BPF classification tool can be successfully used to guide chemical structure activity relationship optimization, enabling antibiotic development and that this approach can be fruitfully applied across species. The BPF classification tool could be potentially applied in primary screening, effectively enabling identification of novel antibacterial compound hits and differentiating their MoA, hence widening the known antibacterial chemical space of existing pharmaceutical compound libraries. More generally, beyond the specific objective of the present work, the proposed approach could be profitably applied to a broader range of diseases amenable to phenotypic drug discovery.
Conflict of interest statement
S.Z., M.V., F.M., A.M., L.W., R.B.M., R.S., K.B., A.I.J., C.L., M.B., K.A. and Ma.P. are full-time employees of F. Hoffmann-La Roche A.G., S.Z., M.V., L.W., Mi.P., H.D., R.S., K.B., A.I.J., C.L., M.B. and K.A. own stocks or stock options in F. Hoffmann-La Roche A.G. C.S.M. is. an employee and holder of stock in Summit Therapeutics plc. T.H., H.H.T. and A.A.R. have no potential conflict of interest.
Figures


References
-
- Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, including tuberculosis., (World Health Organisation, Geneva 2017).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

