Gene Saturation: An Approach to Assess Exploration Stage of Gene Interaction Networks
- PMID: 30899072
- PMCID: PMC6428845
- DOI: 10.1038/s41598-019-41539-w
Gene Saturation: An Approach to Assess Exploration Stage of Gene Interaction Networks
Abstract
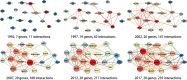
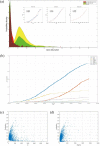
The gene interaction network is one of the most important biological networks and has been studied by many researchers. The gene interaction network provides information about whether the genes in the network can cause or heal diseases. As gene-gene interaction relations are constantly explored, gene interaction networks are evolving. To describe how much a gene has been studied, an approach based on a logistic model for each gene called gene saturation has been proposed, which in most cases, satisfies non-decreasing, correlation and robustness principles. The average saturation of a group of genes can be used to assess the network constructed by these genes. Saturation reflects the distance between known gene interaction networks and the real gene interaction network in a cell. Furthermore, the saturation values of 546 disease gene networks that belong to 15 categories of diseases have been calculated. The disease gene networks' saturation for cancer is significantly higher than that of all other diseases, which means that the disease gene networks' structure for cancer has been more deeply studied than other disease. Gene saturation provides guidance for selecting an experimental subject gene, which may have a large number of unknown interactions.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Robbins KC, Summaria L, Hsieh B, Shah RJ. The peptide chain of human plasmin. mechanism of activation of human plasminogen to plasmin. J. Biol. Chem. 1967;242:2333. - PubMed
-
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 2001;28:21–28. - PubMed
-
- Karopka T, Scheel TS, Glass A. Automatic construction of gene relation networks using text mining and gene expression data. Med. Informatics. 2004;29:169–183. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

