Differences in Systemic IgA Reactivity and Circulating Th Subsets in Healthy Volunteers With Specific Microbiota Enterotypes
- PMID: 30899257
- PMCID: PMC6417458
- DOI: 10.3389/fimmu.2019.00341
Differences in Systemic IgA Reactivity and Circulating Th Subsets in Healthy Volunteers With Specific Microbiota Enterotypes
Abstract
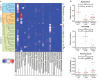
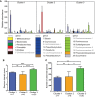
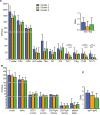
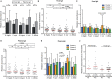
Changes in the intestinal microbiota have been associated with the development of immune-mediated diseases in humans. Additionally, the introduction of defined bacterial species into the mouse intestinal microbiota has been shown to impact on the adaptive immune response. However, how much impact the intestinal microbiota composition actually has on regulating adaptive immunity remains poorly understood. Therefore, we studied aspects of the adaptive immunity in healthy adults possessing distinct intestinal microbiota profiles. The intestinal microbiota composition was determined via Illumina sequencing of bacterial 16S rRNA genes extracted from the feces of 35 individuals. Blood B-cell and T-cell subsets from the same individuals were studied using flow cytometry. Finally, the binding of fecal and plasma Immunoglobulin A (IgA) to intestinal bacteria (associated with health and disease) Bacteroides fragilis, Prevotella copri, Bifidobacterium longum, Clostridium difficile, and Escherichia coli was analyzed using ELISA. Unsupervised clustering of microbiota composition revealed the presence of three clusters within the cohort. Cluster 1 and 2 were similar to previously-described enterotypes with a predominance of Bacteroides in Cluster 1 and Prevotella in Cluster 2. The bacterial diversity (Shannon index) and bacterial richness of Cluster 3 was significantly higher than observed in Clusters 1 and 2, with the Ruminococacceae tending to predominate. Within circulating B- and T-cell subsets, only Th subsets were significantly different between groups of distinct intestinal microbiota. Individuals of Cluster 3 have significantly fewer Th17 and Th22 circulating cells, while Th17.1 cell numbers were increased in individuals of Cluster 1. IgA reactivity to intestinal bacteria was higher in plasma than feces, and individuals of Cluster 1 had significant higher plasma IgA reactivity against B. longum than individuals of Cluster 2. In conclusion, we identified three distinct fecal microbiota clusters, of which two clusters resembled previously-described "enterotypes". Global T-cell and B-cell immunity seemed unaffected, however, circulating Th subsets and plasma IgA reactivity were significantly different between Clusters. Hence, the impact of intestinal bacteria composition on human B cells, T cells and IgA reactivity appears limited in genetically-diverse and environmentally-exposed humans, but can skew antibody reactivity and Th cell subsets.
Keywords: 16S sequencing; IgA reactivity; IgA+ B cells; enterotypes; helper-T cells; intestinal microbiota; γδT cells.
Figures
Similar articles
-
A distinct immunophenotype in children carrying the Blautia enterotype: The Generation R study.Clin Immunol. 2025 Feb;271:110426. doi: 10.1016/j.clim.2025.110426. Epub 2025 Jan 10. Clin Immunol. 2025. PMID: 39800090
-
Analysis of Flagellin-Specific Adaptive Immunity Reveals Links to Dysbiosis in Patients With Inflammatory Bowel Disease.Cell Mol Gastroenterol Hepatol. 2020;9(3):485-506. doi: 10.1016/j.jcmgh.2019.11.012. Epub 2019 Nov 30. Cell Mol Gastroenterol Hepatol. 2020. PMID: 31790809 Free PMC article.
-
Gut microbiota profile in healthy Indonesians.World J Gastroenterol. 2019 Mar 28;25(12):1478-1491. doi: 10.3748/wjg.v25.i12.1478. World J Gastroenterol. 2019. PMID: 30948911 Free PMC article.
-
Immune-microbiota interactions in health and disease.Clin Immunol. 2015 Aug;159(2):122-127. doi: 10.1016/j.clim.2015.05.014. Epub 2015 Jun 30. Clin Immunol. 2015. PMID: 26141651 Free PMC article. Review.
-
Re-thinking the functions of IgA(+) plasma cells.Gut Microbes. 2014;5(5):652-62. doi: 10.4161/19490976.2014.969977. Gut Microbes. 2014. PMID: 25483334 Free PMC article. Review.
Cited by
-
Eosinophil Knockout Humans: Uncovering the Role of Eosinophils Through Eosinophil-Directed Biological Therapies.Annu Rev Immunol. 2021 Apr 26;39:719-757. doi: 10.1146/annurev-immunol-093019-125918. Epub 2021 Mar 1. Annu Rev Immunol. 2021. PMID: 33646859 Free PMC article.
-
Differences in gut microbial patterns associated with salivary biomarkers in young Japanese adults.Biosci Microbiota Food Health. 2020;39(4):243-249. doi: 10.12938/bmfh.2019-034. Epub 2020 Aug 1. Biosci Microbiota Food Health. 2020. PMID: 33117623 Free PMC article.
-
Intestinal microbiota: a new force in cancer immunotherapy.Cell Commun Signal. 2020 Jun 10;18(1):90. doi: 10.1186/s12964-020-00599-6. Cell Commun Signal. 2020. PMID: 32522267 Free PMC article. Review.
-
Oral Microbiome Dysbiosis Is Associated With Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study.Front Microbiol. 2021 Jun 23;12:687513. doi: 10.3389/fmicb.2021.687513. eCollection 2021. Front Microbiol. 2021. PMID: 34248910 Free PMC article.
-
Immunotherapy in Solid Tumors and Gut Microbiota: The Correlation-A Special Reference to Colorectal Cancer.Cancers (Basel). 2020 Dec 25;13(1):43. doi: 10.3390/cancers13010043. Cancers (Basel). 2020. PMID: 33375686 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

