A targeted genomic alteration analysis predicts survival of melanoma patients under BRAF inhibitors
- PMID: 30899440
- PMCID: PMC6422198
- DOI: 10.18632/oncotarget.26707
A targeted genomic alteration analysis predicts survival of melanoma patients under BRAF inhibitors
Abstract
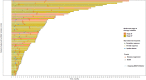
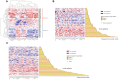
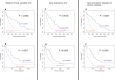
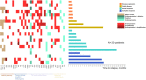
Several mechanisms have been described to elucidate the emergence of resistance to MAPK inhibitors in melanoma and there is a crucial need for biomarkers to identify patients who are likely to achieve a better and long-lasting response to BRAF inhibitors therapy. In this study, we developed a targeted approach combining both mRNA and DNA alterations analysis focusing on relevant gene alterations involved in acquired BRAF inhibitor resistance. We collected baseline tumor samples from 64 melanoma patients at BRAF inhibitor treatment initiation and showed that the presence, prior to treatment, of mRNA over-expression of genes' subset was significantly associated with improved progression free survival and overall survival. The presence of DNA alterations was in favor of better overall survival. The genomic analysis of relapsed-matched tumor samples from 20 patients allowed us to uncover the largest landscape of resistance mechanisms reported to date as at least one resistance mechanism was identified for each patient studied. Alterations in RB1 have been most frequent and hence represent an important additional acquired resistance mechanism. Our targeted genomic analysis emerges as a relevant tool in clinical practice to identify those patients who are more likely to achieve durable response to targeted therapies and to exhaustively describe the spectrum of resistance mechanisms. Our approach can be adapted to new targeted therapies by including newly identified genetic alterations.
Keywords: BRAF inhibitors; melanoma; predictive analysis; targeted genomic alteration; targeted therapy resistance.
Conflict of interest statement
CONFLICTS OF INTEREST BL, JD, CRdM, LG, FJ, AS, MPP, IC, OM, JC, JPF and ND have no conflicts of interest to declare. MB declares a consulting role for Histalim, Bristol-Myers Squibb, and Innate Pharma. SD is a principal investigator in studies conducted by Roche-Genentech and Novartis. CL declares honoraria from Roche, advisory roles at Roche, GSK, Novartis, BMS, MSD, and Amgen and travel accommodation provided by Roche. SM declares a consulting role at Roche, Janssen and Novartis.
Figures



References
-
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Hersey P, Kefford R, Lawrence D, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. - DOI - PMC - PubMed
-
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. - DOI - PMC - PubMed
-
- Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr, Kaempgen E, Martín-Algarra S, Karaszewska B, Mauch C, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. - DOI - PubMed
-
- Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, Chiarion-Sileni V, Lebbe C, Mandalà M, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015;386:444–51. doi: 10.1016/S0140-6736(15)60898-4. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

