Clinical protein science in translational medicine targeting malignant melanoma
- PMID: 30900145
- PMCID: PMC6757020
- DOI: 10.1007/s10565-019-09468-6
Clinical protein science in translational medicine targeting malignant melanoma
Abstract
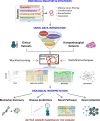
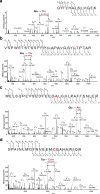
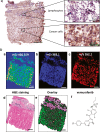
Melanoma of the skin is the sixth most common type of cancer in Europe and accounts for 3.4% of all diagnosed cancers. More alarming is the degree of recurrence that occurs with approximately 20% of patients lethally relapsing following treatment. Malignant melanoma is a highly aggressive skin cancer and metastases rapidly extend to the regional lymph nodes (stage 3) and to distal organs (stage 4). Targeted oncotherapy is one of the standard treatment for progressive stage 4 melanoma, and BRAF inhibitors (e.g. vemurafenib, dabrafenib) combined with MEK inhibitor (e.g. trametinib) can effectively counter BRAFV600E-mutated melanomas. Compared to conventional chemotherapy, targeted BRAFV600E inhibition achieves a significantly higher response rate. After a period of cancer control, however, most responsive patients develop resistance to the therapy and lethal progression. The many underlying factors potentially causing resistance to BRAF inhibitors have been extensively studied. Nevertheless, the remaining unsolved clinical questions necessitate alternative research approaches to address the molecular mechanisms underlying metastatic and treatment-resistant melanoma. In broader terms, proteomics can address clinical questions far beyond the reach of genomics, by measuring, i.e. the relative abundance of protein products, post-translational modifications (PTMs), protein localisation, turnover, protein interactions and protein function. More specifically, proteomic analysis of body fluids and tissues in a given medical and clinical setting can aid in the identification of cancer biomarkers and novel therapeutic targets. Achieving this goal requires the development of a robust and reproducible clinical proteomic platform that encompasses automated biobanking of patient samples, tissue sectioning and histological examination, efficient protein extraction, enzymatic digestion, mass spectrometry-based quantitative protein analysis by label-free or labelling technologies and/or enrichment of peptides with specific PTMs. By combining data from, e.g. phosphoproteomics and acetylomics, the protein expression profiles of different melanoma stages can provide a solid framework for understanding the biology and progression of the disease. When complemented by proteogenomics, customised protein sequence databases generated from patient-specific genomic and transcriptomic data aid in interpreting clinical proteomic biomarker data to provide a deeper and more comprehensive molecular characterisation of cellular functions underlying disease progression. In parallel to a streamlined, patient-centric, clinical proteomic pipeline, mass spectrometry-based imaging can aid in interrogating the spatial distribution of drugs and drug metabolites within tissues at single-cell resolution. These developments are an important advancement in studying drug action and efficacy in vivo and will aid in the development of more effective and safer strategies for the treatment of melanoma. A collaborative effort of gargantuan proportions between academia and healthcare professionals has led to the initiation, establishment and development of a cutting-edge cancer research centre with a specialisation in melanoma and lung cancer. The primary research focus of the European Cancer Moonshot Lund Center is to understand the impact that drugs have on cancer at an individualised and personalised level. Simultaneously, the centre increases awareness of the relentless battle against cancer and attracts global interest in the exceptional research performed at the centre.
Keywords: Cancer moonshot; Clinical proteomics; Malignant melanoma; Post-translational modifications; Translational medicine.
Figures






Similar articles
-
Patient-reported outcomes in patients with resected, high-risk melanoma with BRAFV600E or BRAFV600K mutations treated with adjuvant dabrafenib plus trametinib (COMBI-AD): a randomised, placebo-controlled, phase 3 trial.Lancet Oncol. 2019 May;20(5):701-710. doi: 10.1016/S1470-2045(18)30940-9. Epub 2019 Mar 27. Lancet Oncol. 2019. PMID: 30928620 Clinical Trial.
-
Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial.Lancet Oncol. 2018 Feb;19(2):181-193. doi: 10.1016/S1470-2045(18)30015-9. Epub 2018 Jan 18. Lancet Oncol. 2018. PMID: 29361468 Clinical Trial.
-
Comparison of dabrafenib and trametinib combination therapy with vemurafenib monotherapy on health-related quality of life in patients with unresectable or metastatic cutaneous BRAF Val600-mutation-positive melanoma (COMBI-v): results of a phase 3, open-label, randomised trial.Lancet Oncol. 2015 Oct;16(13):1389-98. doi: 10.1016/S1470-2045(15)00087-X. Lancet Oncol. 2015. PMID: 26433819 Clinical Trial.
-
Current and future roles of targeted therapy and immunotherapy in advanced melanoma.J Manag Care Spec Pharm. 2014 Apr;20(4):346-56. doi: 10.18553/jmcp.2014.20.4.346. J Manag Care Spec Pharm. 2014. PMID: 24684639 Free PMC article. Review.
-
Challenging the heterogeneity of disease presentation in malignant melanoma-impact on patient treatment.Cell Biol Toxicol. 2019 Feb;35(1):1-14. doi: 10.1007/s10565-018-9446-9. Epub 2018 Oct 24. Cell Biol Toxicol. 2019. PMID: 30357519 Free PMC article. Review.
Cited by
-
Digital Barcodes for High-Throughput Screening.Chem Bio Eng. 2024 Jan 26;1(1):2-12. doi: 10.1021/cbe.3c00085. eCollection 2024 Feb 22. Chem Bio Eng. 2024. PMID: 39973970 Free PMC article. Review.
-
A refocus on the advances of single-cell biomedicine.Cell Biol Toxicol. 2020 Oct;36(5):395-398. doi: 10.1007/s10565-020-09551-3. Epub 2020 Aug 10. Cell Biol Toxicol. 2020. PMID: 32779088 Free PMC article. No abstract available.
-
The Hidden Story of Heterogeneous B-raf V600E Mutation Quantitative Protein Expression in Metastatic Melanoma-Association with Clinical Outcome and Tumor Phenotypes.Cancers (Basel). 2019 Dec 9;11(12):1981. doi: 10.3390/cancers11121981. Cancers (Basel). 2019. PMID: 31835364 Free PMC article.
-
Research progress of omics technology in the field of tumor resistance: From single-omics to multi-omics combination application.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021 Jun 28;46(6):620-627. doi: 10.11817/j.issn.1672-7347.2021.200561. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34275931 Free PMC article. Chinese, English.
-
Melanization as unfavorable factor in amelanotic melanoma cell biology.Protoplasma. 2021 Sep;258(5):935-948. doi: 10.1007/s00709-021-01613-5. Epub 2021 Jan 27. Protoplasma. 2021. PMID: 33506271 Free PMC article.
References
-
- Abelin JG, Patel J, Lu X, Feeney CM, Fagbami L, Creech AL, Hu R, Lam D, Davison D, Pino L, Qiao JW, Kuhn E, Officer A, Li J, Abbatiello S, Subramanian A, Sidman R, Snyder E, Carr SA, Jaffe JD. Reduced-representation phosphosignatures measured by quantitative targeted MS capture cellular states and enable large-scale comparison of drug-induced phenotypes. Mol Cell Proteomics. 2016;15:1622–1641. - PMC - PubMed
-
- Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, Costello CE, Cravatt BF, Fenselau C, Garcia BA, Ge Y, Gunawardena J, Hendrickson RC, Hergenrother PJ, Huber CG, Ivanov AR, Jensen ON, Jewett MC, Kelleher NL, Kiessling LL, Krogan NJ, Larsen MR, Loo JA, Ogorzalek Loo RR, Lundberg E, MacCoss MJ, Mallick P, Mootha VK, Mrksich M, Muir TW, Patrie SM, Pesavento JJ, Pitteri SJ, Rodriguez H, Saghatelian A, Sandoval W, Schlüter H, Sechi S, Slavoff SA, Smith LM, Snyder MP, Thomas PM, Uhlén M, van Eyk JE, Vidal M, Walt DR, White FM, Williams ER, Wohlschlager T, Wysocki VH, Yates NA, Young NL, Zhang B. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–214. - PMC - PubMed
-
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, Arachchi H, Arora A, Auman JT, Ayala B, Baboud J, Balasundaram M, Balu S, Barnabas N, Bartlett J, Bartlett P, Bastian BC, Baylin SB, Behera M, Belyaev D, Benz C, Bernard B, Beroukhim R, Bir N, Black AD, Bodenheimer T, Boice L, Boland GM, Bono R, Bootwalla MS, Bosenberg M, Bowen J, Bowlby R, Bristow CA, Brockway-Lunardi L, Brooks D, Brzezinski J, Bshara W, Buda E, Burns WR, Butterfield YSN, Button M, Calderone T, Cappellini GA, Carter C, Carter SL, Cherney L, Cherniack AD, Chevalier A, Chin L, Cho J, Cho RJ, Choi YL, Chu A, Chudamani S, Cibulskis K, Ciriello G, Clarke A, Coons S, Cope L, Crain D, Curley E, Danilova L, D’Atri S, Davidsen T, Davies MA, Delman KA, Demchok JA, Deng QA, Deribe YL, Dhalla N, Dhir R, DiCara D, Dinikin M, Dubina M, Ebrom JS, Egea S, Eley G, Engel J, Eschbacher JM, Fedosenko KV, Felau I, Fennell T, Ferguson ML, Fisher S, Flaherty KT, Frazer S, Frick J, Fulidou V, Gabriel SB, Gao J, Gardner J, Garraway LA, Gastier-Foster JM, Gaudioso C, Gehlenborg N, Genovese G, Gerken M, Gershenwald JE, Getz G, Gomez-Fernandez C, Gribbin T, Grimsby J, Gross B, Guin R, Gutschner T, Hadjipanayis A, Halaban R, Hanf B, Haussler D, Haydu LE, Hayes DN, Hayward NK, Heiman DI, Herbert L, Herman JG, Hersey P, Hoadley KA, Hodis E, Holt RA, Hoon DSB, Hoppough S, Hoyle AP, Huang FW, Huang M, Huang S, Hutter CM, Ibbs M, Iype L, Jacobsen A, Jakrot V, Janning A, Jeck WR, Jefferys SR, Jensen MA, Jones CD, Jones SJM, Ju Z, Kakavand H, Kang H, Kefford RF, Khuri FR, Kim J, Kirkwood JM, Klode J, Korkut A, Korski K, Krauthammer M, Kucherlapati R, Kwong LN, Kycler W, Ladanyi M, Lai PH, Laird PW, Lander E, Lawrence MS, Lazar AJ, Łaźniak R, Lee D, Lee JE, Lee J, Lee K, Lee S, Lee W, Leporowska E, Leraas KM, Li HI, Lichtenberg TM, Lichtenstein L, Lin P, Ling S, Liu J, Liu O, Liu W, Long GV, Lu Y, Ma S, Ma Y, Mackiewicz A, Mahadeshwar HS, Malke J, Mallery D, Manikhas GM, Mann GJ, Marra MA, Matejka B, Mayo M, Mehrabi S, Meng S, Meyerson M, Mieczkowski PA, Miller JP, Miller ML, Mills GB, Moiseenko F, Moore RA, Morris S, Morrison C, Morton D, Moschos S, Mose LE, Muller FL, Mungall AJ, Murawa D, Murawa P, Murray BA, Nezi L, Ng S, Nicholson D, Noble MS, Osunkoya A, Owonikoko TK, Ozenberger BA, Pagani E, Paklina OV, Pantazi A, Parfenov M, Parfitt J, Park PJ, Park WY, Parker JS, Passarelli F, Penny R, Perou CM, Pihl TD, Potapova O, Prieto VG, Protopopov A, Quinn MJ, Radenbaugh A, Rai K, Ramalingam SS, Raman AT, Ramirez NC, Ramirez R, Rao U, Rathmell WK, Ren X, Reynolds SM, Roach J, Robertson AG, Ross MI, Roszik J, Russo G, Saksena G, Saller C, Samuels Y, Sander C, Sander C, Sandusky G, Santoso N, Saul M, Saw RPM, Schadendorf D, Schein JE, Schultz N, Schumacher SE, Schwallier C, Scolyer RA, Seidman J, Sekhar PC, Sekhon HS, Senbabaoglu Y, Seth S, Shannon KF, Sharpe S, Sharpless NE, Shaw KRM, Shelton C, Shelton T, Shen R, Sheth M, Shi Y, Shiau CJ, Shmulevich I, Sica GL, Simons JV, Sinha R, Sipahimalani P, Sofia HJ, Soloway MG, Song X, Sougnez C, Spillane AJ, Spychała A, Stretch JR, Stuart J, Suchorska WM, Sucker A, Sumer SO, Sun Y, Synott M, Tabak B, Tabler TR, Tam A, Tan D, Tang J, Tarnuzzer R, Tarvin K, Tatka H, Taylor BS, Teresiak M, Thiessen N, Thompson JF, Thorne L, Thorsson V, Trent JM, Triche TJ, Jr, Tsai KY, Tsou P, van den Berg DJ, van Allen EM, Veluvolu U, Verhaak RG, Voet D, Voronina O, Walter V, Walton JS, Wan Y, Wang Y, Wang Z, Waring S, Watson IR, Weinhold N, Weinstein JN, Weisenberger DJ, White P, Wilkerson MD, Wilmott JS, Wise L, Wiznerowicz M, Woodman SE, Wu CJ, Wu CC, Wu J, Wu Y, Xi R, Xu AW, Yang D, Yang L, Yang L, Zack TI, Zenklusen JC, Zhang H, Zhang J, Zhang W, Zhao X, Zhu J, Zhu K, Zimmer L, Zmuda E, Zou L. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. - PMC - PubMed
-
- Alonso SR, Tracey L, Ortiz P, Pérez-Gómez B, Palacios J, Pollán M, Linares J, Serrano S, Sáez-Castillo AI, Sánchez L, Pajares R, Sánchez-Aguilera A, Artiga MJ, Piris MA, Rodríguez-Peralto JL. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

