HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles
- PMID: 30902835
- PMCID: PMC6521601
- DOI: 10.1261/rna.069351.118
HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles
Abstract
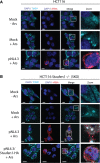
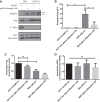
The human immunodeficiency virus type 1 (HIV-1) genomic RNA (vRNA) has two major fates during viral replication: to serve as the template for the major structural and enzymatic proteins, or to be encapsidated and packaged into assembling virions to serve as the genomic vRNA in budding viruses. The dynamic balance between vRNA translation and encapsidation is mediated by numerous host proteins, including Staufen1. During HIV-1 infection, HIV-1 recruits Staufen1 to assemble a distinct ribonucleoprotein complex promoting vRNA encapsidation and viral assembly. Staufen1 also rescues vRNA translation and gene expression during conditions of cellular stress. In this work, we utilized novel Staufen1-/- gene-edited cells to further characterize the contribution of Staufen1 in HIV-1 replication. We observed a marked deficiency in the ability of HIV-1 to dissociate stress granules (SGs) in Staufen1-deficient cells and remarkably, the vRNA repositioned to SGs. These phenotypes were rescued by Staufen1 expression in trans or in cis, but not by a dsRBD-binding mutant, Staufen1F135A. The mistrafficking of the vRNA in these Staufen1-/- cells was also accompanied by a dramatic decrease in viral production and infectivity. This work provides novel insight into the mechanisms by which HIV-1 uses Staufen1 to ensure optimal vRNA translation and trafficking, supporting an integral role for Staufen1 in the HIV-1 life cycle, positioning it as an attractive target for next-generation antiretroviral agents.
Keywords: HIV-1; Staufen1; stress granules; stress response to infection; vRNA trafficking; viral genomic RNA (vRNA).
© 2019 Rao et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Abrahamyan L, Chatel-Chaix L, Ajamian L, Milev M, Monette A, Clément JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, et al. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci 123: 369–383. 10.1242/jcs.055897 - DOI - PubMed
-
- Barajas BC, Tanaka M, Robinson BA, Phuong DJ, Chutiraka K, Reed JC, Lingappa JR. 2018. Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging. PLoS Pathog 14: e1006977 10.1371/journal.ppat.1006977 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
