An Osmoregulatory Mechanism Operating through OmpR and LrhA Controls the Motile-Sessile Switch in the Plant Growth-Promoting Bacterium Pantoea alhagi
- PMID: 30902852
- PMCID: PMC6498166
- DOI: 10.1128/AEM.00077-19
An Osmoregulatory Mechanism Operating through OmpR and LrhA Controls the Motile-Sessile Switch in the Plant Growth-Promoting Bacterium Pantoea alhagi
Abstract
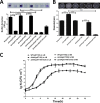
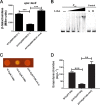
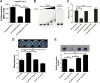
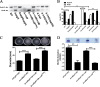
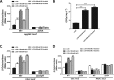
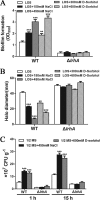
Adaptation to osmotic stress is crucial for bacterial growth and survival in changing environments. Although a large number of osmotic stress response genes have been identified in various bacterial species, how osmotic changes affect bacterial motility, biofilm formation, and colonization of host niches remains largely unknown. In this study, we report that the LrhA regulator is an osmoregulated transcription factor that directly binds to the promoters of the flhDC, eps, and opgGH operons and differentially regulates their expression, thus inhibiting motility and promoting exopolysaccharide (EPS) production, synthesis of osmoregulated periplasmic glucans (OPGs), biofilm formation, and root colonization of the plant growth-promoting bacterium Pantoea alhagi LTYR-11Z. Further, we observed that the LrhA-regulated OPGs control RcsCD-RcsB activation in a concentration-dependent manner, and a high concentration of OPGs induced by increased medium osmolarity is maintained to achieve the high level of activation of the Rcs phosphorelay, which results in enhanced EPS synthesis and decreased motility in P. alhagi Moreover, we showed that the osmosensing regulator OmpR directly binds to the promoter of lrhA and promotes its expression, while lrhA expression is feedback inhibited by the activated Rcs phosphorelay system. Overall, our data support a model whereby P. alhagi senses environmental osmolarity changes through the EnvZ-OmpR two-component system and LrhA to regulate the synthesis of OPGs, EPS production, and flagellum-dependent motility, thereby employing a hierarchical signaling cascade to control the transition between a motile lifestyle and a biofilm lifestyle.IMPORTANCE Many motile bacterial populations form surface-attached biofilms in response to specific environmental cues, including osmotic stress in a range of natural and host-related systems. However, cross talk between bacterial osmosensing, swimming, and biofilm formation regulatory networks is not fully understood. Here, we report that the pleiotropic regulator LrhA in Pantoea alhagi is involved in the regulation of flagellar motility, biofilm formation, and host colonization and responds to osmotic upshift. We further show that this sensing relies on the EnvZ-OmpR two-component system that was known to detect changes in external osmotic stress. The EnvZ-OmpR-LrhA osmosensing signal transduction cascade is proposed to increase bacterial fitness under hyperosmotic conditions inside the host. Our work proposes a novel regulatory mechanism that links osmosensing and motile-sessile lifestyle transitions, which may provide new approaches to prevent or promote the formation of biofilms and host colonization in P. alhagi and other bacteria possessing a similar osmoregulatory mechanism.
Keywords: LrhA; OPGs; OmpR; Rcs phosphorelay; biofilm; exopolysaccharides; motility; osmosensing.
Copyright © 2019 American Society for Microbiology.
Figures


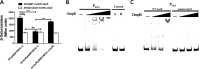

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

