Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities
- PMID: 30902854
- PMCID: PMC6498177
- DOI: 10.1128/AEM.00110-19
Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities
Abstract
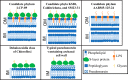
Recent experimental and bioinformatic advances enable the recovery of genomes belonging to yet-uncultured microbial lineages directly from environmental samples. Here, we report on the recovery and characterization of single amplified genomes (SAGs) and metagenome-assembled genomes (MAGs) representing candidate phylum LCP-89, previously defined based on 16S rRNA gene sequences. Analysis of LCP-89 genomes recovered from Zodletone Spring, an anoxic spring in Oklahoma, predicts slow-growing, rod-shaped organisms. LCP-89 genomes contain genes for cell wall lipopolysaccharide (LPS) production but lack the entire machinery for peptidoglycan biosynthesis, suggesting an atypical cell wall structure. The genomes, however, encode S-layer homology domain-containing proteins, as well as machinery for the biosynthesis of CMP-legionaminate, inferring the possession of an S-layer glycoprotein. A nearly complete chemotaxis machinery coupled to the absence of flagellar synthesis and assembly genes argues for the utilization of alternative types of motility. A strict anaerobic lifestyle is predicted, with dual respiratory (nitrite ammonification) and fermentative capacities. Predicted substrates include a wide range of sugars and sugar alcohols and a few amino acids. The capability of rhamnose metabolism is confirmed by the identification of bacterial microcompartment genes to sequester the toxic intermediates generated. Comparative genomic analysis identified differences in oxygen sensitivities, respiratory capabilities, substrate utilization preferences, and fermentation end products between LCP-89 genomes and those belonging to its four sister phyla (Calditrichota, SM32-31, AABM5-125-24, and KSB1) within the broader FCB (Fibrobacteres-Chlorobi-Bacteroidetes) superphylum. Our results provide a detailed characterization of members of the candidate division LCP-89 and highlight the importance of reconciling 16S rRNA-based and genome-based phylogenies.IMPORTANCE Our understanding of the metabolic capacities, physiological preferences, and ecological roles of yet-uncultured microbial phyla is expanding rapidly. Two distinct approaches are currently being utilized for characterizing microbial communities in nature: amplicon-based 16S rRNA gene surveys for community characterization and metagenomics/single-cell genomics for detailed metabolic reconstruction. The occurrence of multiple yet-uncultured bacterial phyla has been documented using 16S rRNA surveys, and obtaining genome representatives of these yet-uncultured lineages is critical to our understanding of the role of yet-uncultured organisms in nature. This study provides a genomics-based analysis highlighting the structural features and metabolic capacities of a yet-uncultured bacterial phylum (LCP-89) previously identified in 16S rRNA surveys for which no prior genomes have been described. Our analysis identifies several interesting structural features for members of this phylum, e.g., lack of peptidoglycan biosynthetic machinery and the ability to form bacterial microcompartments. Predicted metabolic capabilities include degradation of a wide range of sugars, anaerobic respiratory capacity, and fermentative capacities. In addition to the detailed structural and metabolic analysis provided for candidate division LCP-89, this effort represents an additional step toward a unified scheme for microbial taxonomy by reconciling 16S rRNA gene-based and genomics-based taxonomic outlines.
Keywords: candidate phyla; environmental genomics; metagenomic bins; single-cell genomics.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, The Earth Microbiome Project Consortium. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. - DOI - PMC - PubMed

