Determinants of Phage Host Range in Staphylococcus Species
- PMID: 30902858
- PMCID: PMC6532042
- DOI: 10.1128/AEM.00209-19
Determinants of Phage Host Range in Staphylococcus Species
Abstract
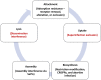
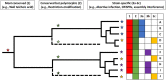
Bacteria in the genus Staphylococcus are important targets for phage therapy due to their prevalence as pathogens and increasing antibiotic resistance. Here we review Staphylococcus outer surface features and specific phage resistance mechanisms that define the host range, the set of strains that an individual phage can potentially infect. Phage infection goes through five distinct phases: attachment, uptake, biosynthesis, assembly, and lysis. Adsorption inhibition, encompassing outer surface teichoic acid receptor alteration, elimination, or occlusion, limits successful phage attachment and entry. Restriction-modification systems (in particular, type I and IV systems), which target phage DNA inside the cell, serve as the major barriers to biosynthesis as well as transduction and horizontal gene transfer between clonal complexes and species. Resistance to late stages of infection occurs through mechanisms such as assembly interference, in which staphylococcal pathogenicity islands siphon away superinfecting phage proteins to package their own DNA. While genes responsible for teichoic acid biosynthesis, capsule, and restriction-modification are found in most Staphylococcus strains, a variety of other host range determinants (e.g., clustered regularly interspaced short palindromic repeats, abortive infection, and superinfection immunity) are sporadic. The fitness costs of phage resistance through teichoic acid structure alteration could make staphylococcal phage therapies promising, but host range prediction is complex because of the large number of genes involved, and the roles of many of these are unknown. In addition, little is known about the genetic determinants that contribute to host range expansion in the phages themselves. Future research must identify host range determinants, characterize resistance development during infection and treatment, and examine population-wide genetic background effects on resistance selection.
Keywords: CRISPR; host range; phage resistance; phage therapy; staphylococci.
Copyright © 2019 Moller et al.
Figures

References
-
- Young R, Bläsi U. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev 17:195–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

