Biofilm-Constructing Variants of Paraburkholderia phytofirmans PsJN Outcompete the Wild-Type Form in Free-Living and Static Conditions but Not In Planta
- PMID: 30902863
- PMCID: PMC6532049
- DOI: 10.1128/AEM.02670-18
Biofilm-Constructing Variants of Paraburkholderia phytofirmans PsJN Outcompete the Wild-Type Form in Free-Living and Static Conditions but Not In Planta
Abstract
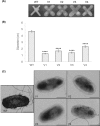
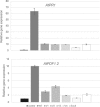
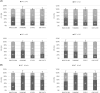
Members of the genus Burkholderia colonize diverse ecological niches. Among the plant-associated strains, Paraburkholderia phytofirmans PsJN is an endophyte with a broad host range. In a spatially structured environment (unshaken broth cultures), biofilm-constructing specialists of P. phytofirmans PsJN colonizing the air-liquid interface arose at high frequency. In addition to forming a robust biofilm in vitro and in planta on Arabidopsis roots, those mucoid phenotypic variants display a reduced swimming ability and modulate the expression of several microbe-associated molecular patterns (MAMPs), including exopolysaccharides (EPS), flagellin, and GroEL. Interestingly, the variants induce low PR1 and PDF1.2 expression compared to that of the parental strain, suggesting a possible evasion of plant host immunity. We further demonstrated that switching from the planktonic to the sessile form did not involve quorum-sensing genes but arose from spontaneous mutations in two genes belonging to an iron-sulfur cluster: hscA (encoding a cochaperone protein) and iscS (encoding a cysteine desulfurase). A mutational approach validated the implication of these two genes in the appearance of variants. We showed for the first time that in a heterogeneous environment, P. phytofirmans strain PsJN is able to rapidly diversify and coexpress a variant that outcompete the wild-type form in free-living and static conditions but not in plantaIMPORTANCEParaburkholderia phytofirmans strain PsJN is a well-studied plant-associated bacterium known to induce resistance against biotic and abiotic stresses. In this work, we described the spontaneous appearance of mucoid variants in PsJN from static cultures. We showed that the conversion from the wild-type (WT) form to variants (V) correlates with an overproduction of EPS, an enhanced ability to form biofilm in vitro and in planta, and a reduced swimming motility. Our results revealed also that these phenotypes are in part associated with spontaneous mutations in an iron-sulfur cluster. Overall, the data provided here allow a better understanding of the adaptive mechanisms likely developed by P. phytofirmans PsJN in a heterogeneous environment.
Keywords: Paraburkholderia phytofirmans; biofilm; competition; iron-sulfur cluster; plant defense; static cultures.
Copyright © 2019 American Society for Microbiology.
Figures


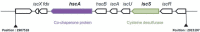


References
-
- Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, Hauser L, Córdova M, Gómez L, González M, Land M, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx CJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IB, Tiedje JM. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci U S A 103:15280–15287. doi: 10.1073/pnas.0606924103. - DOI - PMC - PubMed
-
- Sawana A, Adeolu M, Gupta RS. 2014. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429. doi: 10.3389/fgene.2014.00429. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

