Sterol regulatory element binding protein 1 couples mechanical cues and lipid metabolism
- PMID: 30902980
- PMCID: PMC6430766
- DOI: 10.1038/s41467-019-09152-7
Sterol regulatory element binding protein 1 couples mechanical cues and lipid metabolism
Abstract
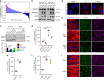
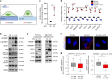
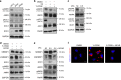
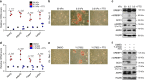
Sterol regulatory element binding proteins (SREBPs) are a family of transcription factors that regulate lipid biosynthesis and adipogenesis by controlling the expression of several enzymes required for cholesterol, fatty acid, triacylglycerol and phospholipid synthesis. In vertebrates, SREBP activation is mainly controlled by a complex and well-characterized feedback mechanism mediated by cholesterol, a crucial bio-product of the SREBP-activated mevalonate pathway. In this work, we identified acto-myosin contractility and mechanical forces imposed by the extracellular matrix (ECM) as SREBP1 regulators. SREBP1 control by mechanical cues depends on geranylgeranyl pyrophosphate, another key bio-product of the mevalonate pathway, and impacts on stem cell fate in mouse and on fat storage in Drosophila. Mechanistically, we show that activation of AMP-activated protein kinase (AMPK) by ECM stiffening and geranylgeranylated RhoA-dependent acto-myosin contraction inhibits SREBP1 activation. Our results unveil an unpredicted and evolutionary conserved role of SREBP1 in rewiring cell metabolism in response to mechanical cues.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

