High levels of PIWI-interacting RNAs are present in the small RNA landscape of prostate epithelium from vitamin D clinical trial specimens
- PMID: 30905091
- PMCID: PMC6593815
- DOI: 10.1002/pros.23789
High levels of PIWI-interacting RNAs are present in the small RNA landscape of prostate epithelium from vitamin D clinical trial specimens
Abstract
Background: Vitamin D, a hormone that acts through the nuclear vitamin D receptor (VDR), upregulates antitumorigenic microRNA in prostate epithelium. This may contribute to the lower levels of aggressive prostate cancer (PCa) observed in patients with high serum vitamin D. The small noncoding RNA (ncRNA) landscape includes many other RNA species that remain uncharacterized in prostate epithelium and their potential regulation by vitamin D is unknown.
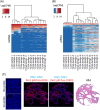
Methods: Laser capture microdissection (LCM) followed by small-RNA sequencing was used to identify ncRNAs in the prostate epithelium of tissues from a vitamin D-supplementation trial. VDR chromatin immunoprecipitation-sequencing was performed to identify vitamin D genomic targets in primary prostate epithelial cells.
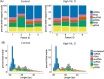
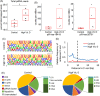
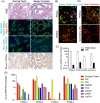
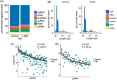
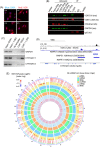
Results: Isolation of epithelium by LCM increased sample homogeneity and captured more diversity in ncRNA species compared with publicly available small-RNA sequencing data from benign whole prostate. An abundance of PIWI-interacting RNAs (piRNAs) was detected in normal prostate epithelium. The obligate binding partners of piRNAs, PIWI-like (PIWIL) proteins, were also detected in prostate epithelium. High prostatic vitamin D levels were associated with increased expression of piRNAs. VDR binding sites were located near several ncRNA biogenesis genes and genes regulating translation and differentiation.
Conclusions: Benign prostate epithelium expresses both piRNA and PIWIL proteins, suggesting that these small ncRNA may serve an unknown function in the prostate. Vitamin D may increase the expression of prostatic piRNAs. VDR binding sites in primary prostate epithelial cells are consistent with its reported antitumorigenic functions and a role in ncRNA biogenesis.
Keywords: ChIP-sequencing; PIWI-interacting RNA; prostate; small-RNA sequencing; vitamin D.
© 2019 The Authors. The Prostate Published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures

Similar articles
-
Differential expression and regulation of vitamin D hydroxylases and inflammatory genes in prostate stroma and epithelium by 1,25-dihydroxyvitamin D in men with prostate cancer and an in vitro model.J Steroid Biochem Mol Biol. 2015 Apr;148:156-65. doi: 10.1016/j.jsbmb.2014.10.004. Epub 2014 Oct 8. J Steroid Biochem Mol Biol. 2015. PMID: 25305352 Free PMC article. Clinical Trial.
-
microRNAs and DICER1 are regulated by 1,25-dihydroxyvitamin D in prostate stroma.J Steroid Biochem Mol Biol. 2017 Mar;167:192-202. doi: 10.1016/j.jsbmb.2017.01.004. Epub 2017 Jan 12. J Steroid Biochem Mol Biol. 2017. PMID: 28089917 Free PMC article. Clinical Trial.
-
PIWIL-2 and piRNAs are regularly expressed in epithelia of the skin and their expression is related to differentiation.Arch Dermatol Res. 2020 Dec;312(10):705-714. doi: 10.1007/s00403-020-02052-7. Epub 2020 Mar 12. Arch Dermatol Res. 2020. PMID: 32166374 Free PMC article.
-
The biogenesis and biological function of PIWI-interacting RNA in cancer.J Hematol Oncol. 2021 Jun 12;14(1):93. doi: 10.1186/s13045-021-01104-3. J Hematol Oncol. 2021. PMID: 34118972 Free PMC article. Review.
-
Vitamin D in Prostate Cancer.Vitam Horm. 2016;100:321-55. doi: 10.1016/bs.vh.2015.10.012. Epub 2015 Dec 8. Vitam Horm. 2016. PMID: 26827958 Review.
Cited by
-
Genomic Insights into Non-steroidal Nuclear Receptors in Prostate and Breast Cancer.Adv Exp Med Biol. 2022;1390:227-239. doi: 10.1007/978-3-031-11836-4_13. Adv Exp Med Biol. 2022. PMID: 36107322
-
PIWI family proteins as prognostic markers in cancer: a systematic review and meta-analysis.Cell Mol Life Sci. 2020 Jun;77(12):2289-2314. doi: 10.1007/s00018-019-03403-y. Epub 2019 Dec 9. Cell Mol Life Sci. 2020. PMID: 31814070 Free PMC article.
-
Vitamin D sufficiency enhances differentiation of patient-derived prostate epithelial organoids.iScience. 2021 Jan 5;24(1):101974. doi: 10.1016/j.isci.2020.101974. eCollection 2021 Jan 22. iScience. 2021. PMID: 33458620 Free PMC article.
-
Piwi-interacting RNAs play a role in vitamin C-mediated effects on endothelial aging.Int J Med Sci. 2020 Mar 26;17(7):946-952. doi: 10.7150/ijms.42586. eCollection 2020. Int J Med Sci. 2020. PMID: 32308548 Free PMC article.
-
Expression of collagen-related piRNA is dysregulated in cultured dermal fibroblasts derived from patients with scleroderma.Intractable Rare Dis Res. 2023 Nov;12(4):241-245. doi: 10.5582/irdr.2023.01056. Intractable Rare Dis Res. 2023. PMID: 38024581 Free PMC article.
References
-
- Forrest KYZ, Stuhldreher WL. Prevalence and correlates of vitamin D deficiency in US adults. Nutr Res. 2011;31(1):48‐54. - PubMed
-
- Daraghmeh AH, Bertoia ML, Al‐Qadi MO, Abdulbaki AM, Roberts MB, Eaton CB. Evidence for the vitamin D hypothesis: the NHANES III extended mortality follow‐up. Atherosclerosis. 2016;255:96‐101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

