Disease Heritability Enrichment of Regulatory Elements Is Concentrated in Elements with Ancient Sequence Age and Conserved Function across Species
- PMID: 30905396
- PMCID: PMC6451699
- DOI: 10.1016/j.ajhg.2019.02.008
Disease Heritability Enrichment of Regulatory Elements Is Concentrated in Elements with Ancient Sequence Age and Conserved Function across Species
Abstract
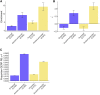
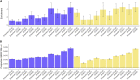
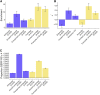
Regulatory elements, e.g., enhancers and promoters, have been widely reported to be enriched for disease and complex trait heritability. We investigated how this enrichment varies with the age of the underlying genome sequence, the conservation of regulatory function across species, and the target gene of the regulatory element. We estimated heritability enrichment by applying stratified LD score regression to summary statistics from 41 independent diseases and complex traits (average N = 320K) and meta-analyzing results across traits. Enrichment of human putative enhancers and promoters was larger in elements with older sequence age, assessed via alignment with other species irrespective of conserved functionality: putative enhancer elements with ancient sequence age (older than the split between marsupial and placental mammals) were 8.8× enriched (versus 2.5× for all putative enhancers; p = 3e-14), and promoter elements with ancient sequence age were 13.5× enriched (versus 5.1× for all promoters; p = 5e-16). Enrichment of human putative enhancers and promoters was also larger in elements whose regulatory function was conserved across species, e.g., human putative enhancers that were enhancers in ≥5 of 9 other mammals were 4.6× enriched (p = 5e-12 versus all putative enhancers). Enrichment of human promoters was larger in promoters of loss-of-function intolerant genes: 12.0× enrichment (p = 8e-15 versus all promoters). The mean value of several measures of negative selection within these genomic annotations mirrored all of these findings. Notably, the annotations with these excess heritability enrichments were jointly significant conditional on each other and on our baseline-LD model, which includes a broad set of coding, conserved, regulatory, and LD-related annotations.
Keywords: enhancer; genetic architecture; heritability; promoter; regulatory elements.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

