Mesenchymal Niche-Specific Expression of Cxcl12 Controls Quiescence of Treatment-Resistant Leukemia Stem Cells
- PMID: 30905620
- PMCID: PMC6499704
- DOI: 10.1016/j.stem.2019.02.018
Mesenchymal Niche-Specific Expression of Cxcl12 Controls Quiescence of Treatment-Resistant Leukemia Stem Cells
Erratum in
-
Mesenchymal Niche-Specific Expression of Cxcl12 Controls Quiescence of Treatment-Resistant Leukemia Stem Cells.Cell Stem Cell. 2020 Jan 2;26(1):123. doi: 10.1016/j.stem.2019.11.013. Cell Stem Cell. 2020. PMID: 31901250 Free PMC article. No abstract available.
Abstract
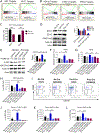
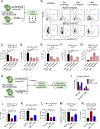
Chronic myeloid leukemia (CML) originates in a hematopoietic stem cell (HSC) transformed by the breakpoint cluster region (BCR)-abelson (ABL) oncogene and is effectively treated with tyrosine kinase inhibitors (TKIs). TKIs do not eliminate disease-propagating leukemic stem cells (LSCs), suggesting a deeper understanding of niche-dependent regulation of CML LSCs is required to eradicate disease. Cxcl12 is expressed in bone marrow niches and controls HSC maintenance, and here, we show that targeted deletion of Cxcl12 from mesenchymal stromal cells (MSCs) reduces normal HSC numbers but promotes LSC expansion by increasing self-renewing cell divisions, possibly through enhanced Ezh2 activity. In contrast, endothelial cell-specific Cxcl12 deletion decreases LSC proliferation, suggesting niche-specific effects. During CML development, abnormal clusters of colocalized MSCs and LSCs form but disappear upon Cxcl12 deletion. Moreover, MSC-specific deletion of Cxcl12 increases LSC elimination by TKI treatment. These findings highlight a critical role of niche-specific effects of Cxcl12 expression in maintaining quiescence of TKI-resistant LSC populations.
Keywords: CXCL12; TKI; bone marrow microenvironment; chronic myelogenous leukemia; drug resistance; hematopoietic stem cells; leukemia stem cells; mesenchymal stromal cells.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing financial interests.
Figures





Comment in
-
Tumor Cell Dormancy-Triggered by the Niche.Dev Cell. 2019 May 6;49(3):311-312. doi: 10.1016/j.devcel.2019.04.022. Dev Cell. 2019. PMID: 31063749
References
-
- Abe-Suzuki S, Kurata M, Abe S, Onishi I, Kirimura S, Nashimoto M, Murayama T, Hidaka M, and Kitagawa M (2014). CXCL12+ stromal cells as bone marrow niche for CD34+ hematopoietic cells and their association with disease progression in myelodysplastic syndromes. Laboratory investigation; a journal of technical methods and pathology 94, 1212–1223. - PubMed
-
- Abraham M, Klein S, Bulvik B, Wald H, Weiss ID, Olam D, Weiss L, Beider K, Eizenberg O, Wald O, et al. (2017). The CXCR4 inhibitor BL-8040 induces the apoptosis of AML blasts by downregulating ERK, BCL-2, MCL-1 and cyclin-D1 via altered miR-15a/16–1 expression. Leukemia - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

