Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma
- PMID: 30906654
- PMCID: PMC6422384
- DOI: 10.1080/2162402X.2018.1561106
Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma
Abstract
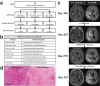
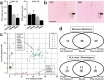
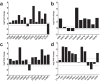
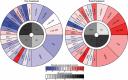
Neoantigens represent promising targets for personalized cancer vaccine strategies. However, the feasibility of this approach in lower mutational burden tumors like glioblastoma (GBM) remains unknown. We have previously reported the use of an immunogenomics pipeline to identify candidate neoantigens in preclinical models of GBM. Here, we report the application of the same immunogenomics pipeline to identify candidate neoantigens and guide screening for neoantigen-specific T cell responses in a patient with GBM treated with a personalized synthetic long peptide vaccine following autologous tumor lysate DC vaccination. Following vaccination, reactivity to three HLA class I- and five HLA class II-restricted candidate neoantigens were detected by IFN-γ ELISPOT in peripheral blood. A similar pattern of reactivity was observed among isolated post-treatment tumor-infiltrating lymphocytes. Genomic analysis of pre- and post-treatment GBM reflected clonal remodeling. These data demonstrate the feasibility and translational potential of a therapeutic neoantigen-based vaccine approach in patients with primary CNS tumors.
Keywords: Neoantigen; clonal evolution; glioblastoma; immunogenomics; personalized vaccine.
Figures


References
-
- Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie W-R, Hildebrand WH, Mardis ER, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi:10.1126/science.aaa3828. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
