Codweb: Whole-genome sequencing uncovers extensive reticulations fueling adaptation among Atlantic, Arctic, and Pacific gadids
- PMID: 30906856
- PMCID: PMC6426462
- DOI: 10.1126/sciadv.aat8788
Codweb: Whole-genome sequencing uncovers extensive reticulations fueling adaptation among Atlantic, Arctic, and Pacific gadids
Abstract
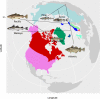
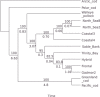
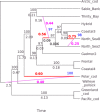
Introgressive hybridization creates networks of genetic relationships across species. Among marine fish of the Gadidae family, Pacific cod and walleye pollock are separate invasions of an Atlantic cod ancestor into the Pacific. Cods are ecological success stories, and their ecologies allow them to support the largest fisheries of the world. The enigmatic walleye pollock differs morphologically, behaviorally, and ecologically from its relatives, representing a niche shift. Here, we apply whole-genome sequencing to Pacific, Arctic, and Atlantic gadids and reveal extensive introgression among them with the ABBA-BABA test and pseudolikelihood phylogenetic network analysis. We propose that walleye pollock resulted from extensive adaptive introgression or homoploid hybrid speciation. The path of evolution of these taxa is more web than a tree. Their ability to invade and expand into new habitats and become ecologically successful may depend on genes acquired through adaptive introgression or hybrid speciation.
Figures



References
-
- Clark A. G., Messer P. W., Conundrum of jumbled mosquito genomes. Science 347, 27–28 (2015). - PubMed
-
- Mallet J., Hybrid speciation. Nature 446, 279–283 (2007). - PubMed
-
- Rieseberg L. H., Hybrid origin of plant species. Annu. Rev. Ecol. Syst. 28, 359–389 (1997).
-
- Mavárez J., Salazar C. A., Bermingham E., Salcedo C., Jiggins C. D., Linares M., Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871 (2006). - PubMed
-
- Cui R., Schumer M., Kruesi K., Walter R., Andolfatto P., Rosenthal G. G., Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 67, 2166–2179 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

